Human MATE1 (BI3323-Aug2025)
From Proteopedia
| Line 24: | Line 24: | ||
cimetidine reduces metformin clearance by competitive inhibition. | cimetidine reduces metformin clearance by competitive inhibition. | ||
| - | [[Image: | + | [[Image:Figure3 ligand view.png]] |
== Relevance == | == Relevance == | ||
Revision as of 17:32, 30 November 2025
Contents |
Introduction
Human MATE1 (multidrug and toxin extrusion protein 1) is a critical membrane transporter expressed in the kidney and liver, where it mediates the final step in removing a broad range of drugs and metabolites from the body through urine and bile. MATE1 is the primary transporter responsible for clearing clinically important drugs such as metformin, an antidiabetic drug used by millions of patients worldwide, and plays a crucial role in drug pharmacokinetics and therapeutic efficacy. Understanding how MATE1 recognizes and binds different substrates is essential for predicting drug-drug interactions and designing new therapeutic agents that can be safely administered without accumulating to toxic levels.
Structure and methods
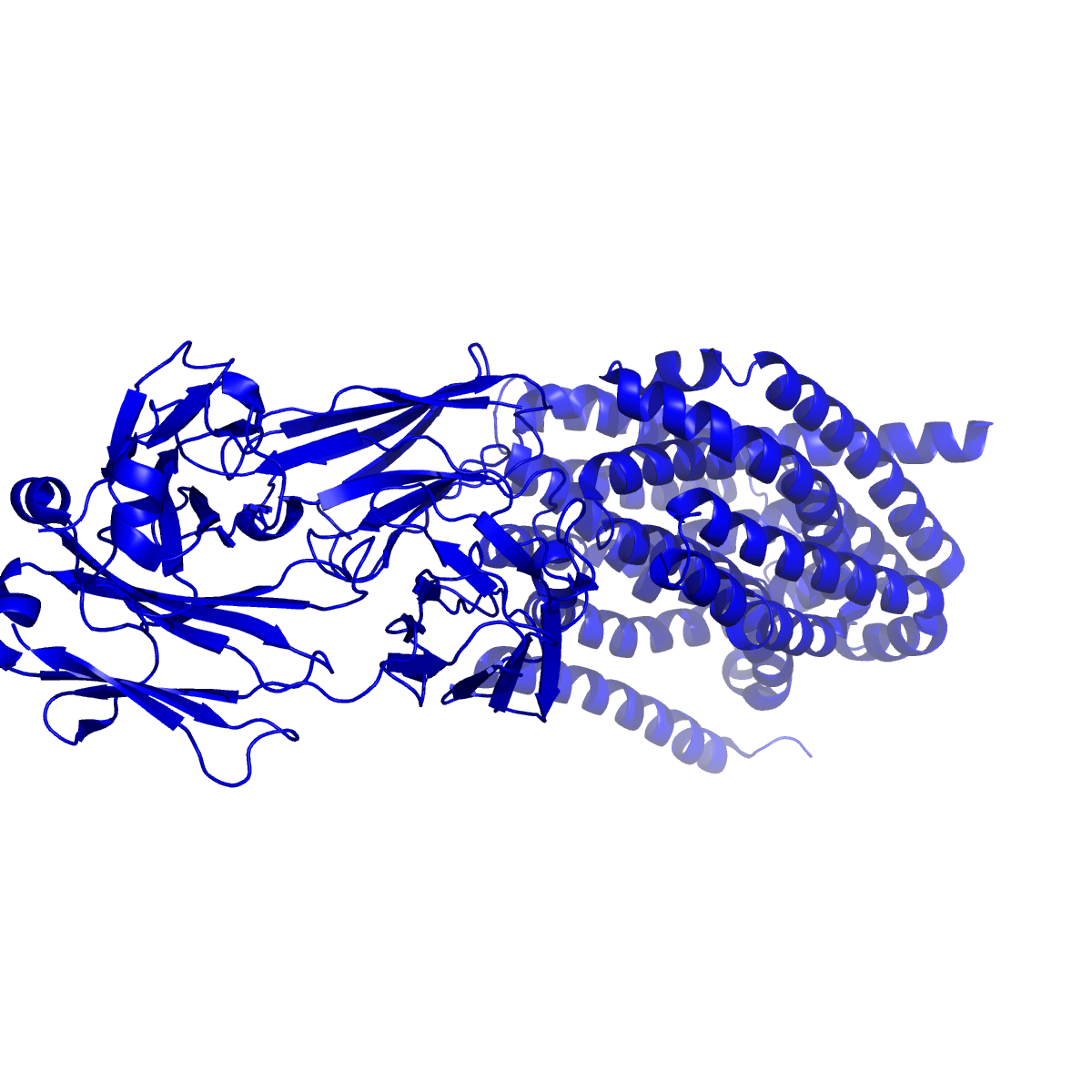
The structure of hMATE1 was first determined using cryo-electron microscopy (cryo-EM), a cutting-edge technique allowing visualization of proteins at near-atomic resolution, to determine the three-dimensional structures of hMATE1 in complex with two different substrates, metformin and MPP, and one inhibitor, cimetidine. The resolution obtained for hMATE1 complexes was 2.3 Å for MATE1-metformin, 3.1 Å for MATE1-MPP (a model organic cation), and 3.3 Å for MATE1-cimetidine (an H2-receptor antagonist that blocks MATE1). To increase protein stability for structure determination, engineered antibody fragments (Fabs) that recognized the transporter were used. Complementary molecular dynamics simulations and functional validation studies with site-directed mutants confirmed the structural findings.
Structural highlights
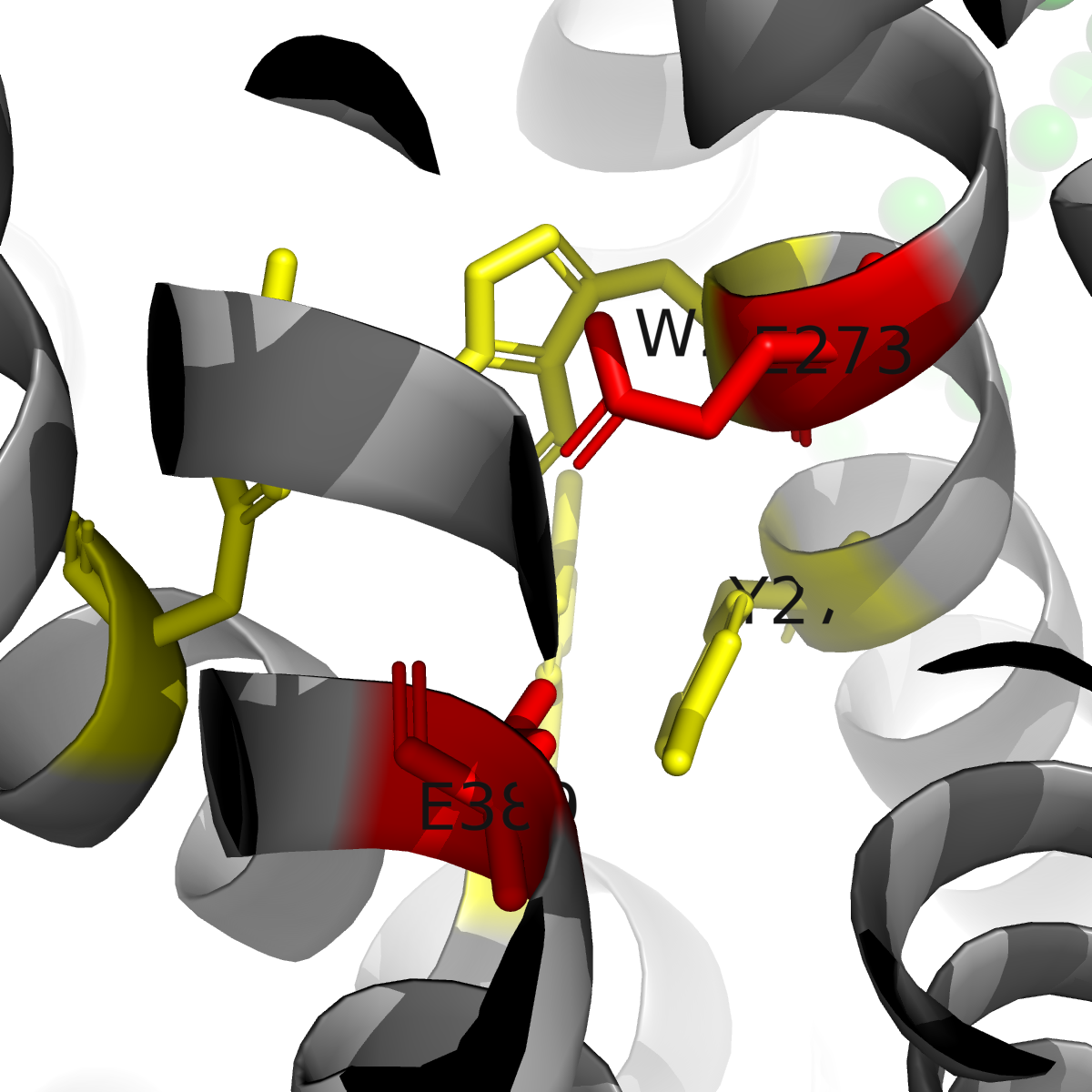
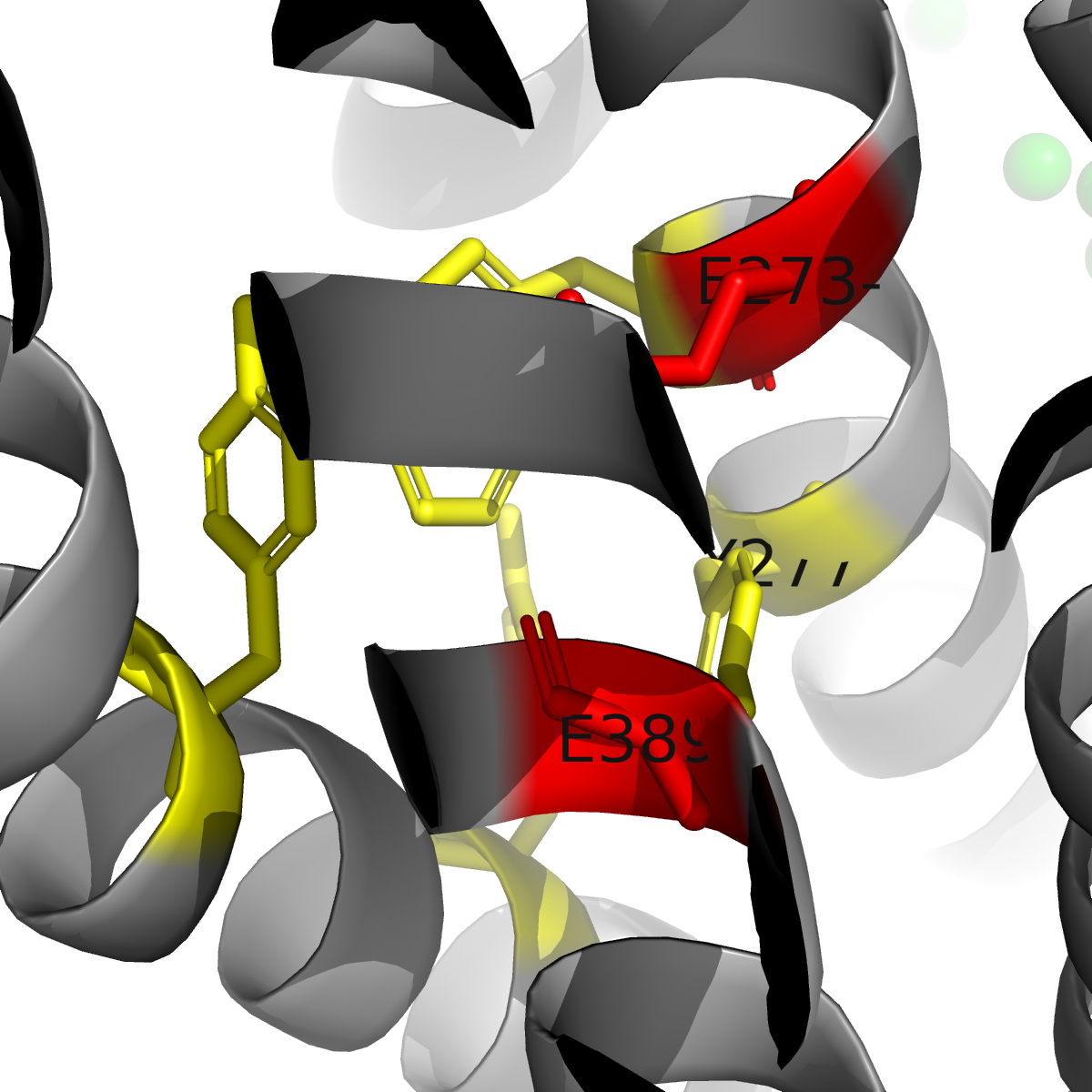
The structures reveal that hMATE1's substrate binding pocket is located within the C-terminal lobe of the transporter, embedded halfway through the membrane. This pocket is lined by two critical glutamate residues (E273 and E389) that create a negatively charged environment, essential for accommodating cationic (positively charged) drug substrates. Additional aromatic residues—W274, Y277, Y299, and Y306—form stabilizing interactions including cation-π and π-π stacking interactions with different substrates. Different ligands show dramatically different binding modes, including the three ligands in the study. Metformin exhibits high mobility within the pocket, interacting through Coulomb interactions with the glutamates and cation-π interactions with aromatic residues. In contrast, cimetidine shows markedly different binding geometry, with its methyl-imidazole ring "locked" between Y277 and Y306 through π-π stacking, forming a hydrogen bond with E389. This more stable binding geometry explains cimetidine's significantly higher inhibitory potency (IC50 = 2.7 µM) compared to metformin (IC50 = 58 µM), directly accounting for the clinically observed drug-drug interaction where cimetidine reduces metformin clearance by competitive inhibition.
Relevance
These structures provide the first atomic-level understanding of human MATE transporter substrate recognition, resolving earlier uncertainty about whether the binding pocket was located in the N-lobe or C-lobe. This knowledge is crucial for predicting which new drug candidates might interact with hMATE1, information that is now required by regulatory agencies (FDA and EMA) during drug development. The findings demonstrate the molecular basis of MATE1's ability to transport structurally diverse compounds and explain how genetic variations or drug-induced inhibition of MATE1 can lead to altered drug clearance.
References
Romane, K., Peteani, G., Mukherjee, S. et al. Structural basis of drug recognition by human MATE1 transporter. Nat Commun 16, 9444 (2025). https://doi.org/10.1038/s41467-025-64490-z