Sandbox Aryan 20221057 BI3323-Aug2025
From Proteopedia
| Line 3: | Line 3: | ||
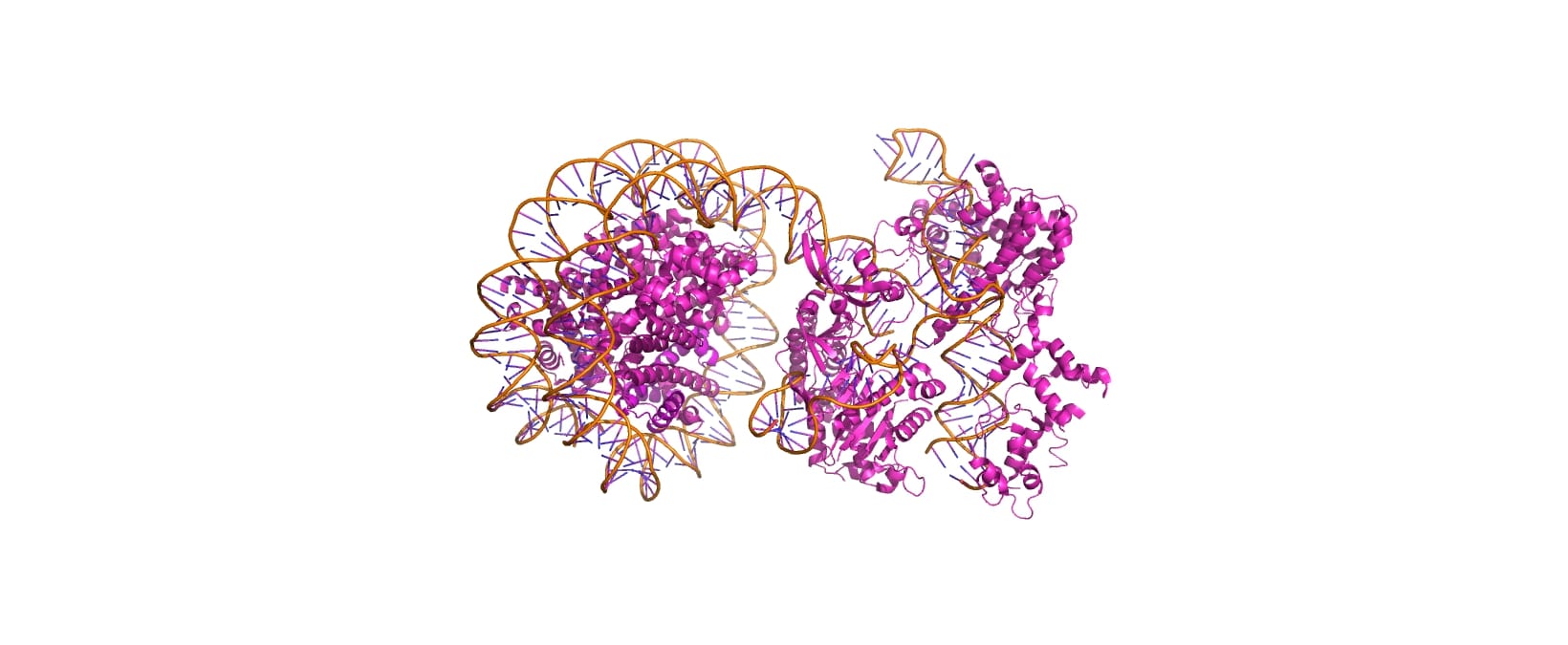
Cas9-sgRNA ribonucleoprotein targets linker DNA (PAM1/PAM28) and entry-exit regions (SHL6) of nucleosomes, avoiding tightly wrapped core DNA (SHL0-5), as revealed by native-PAGE on Widom 601 nucleosomes. The cryo-EM structure (PDB: 8YNY, 4.52 Å, EMDB: EMD-39431) captures the post-cleavage ternary complex at PAM1, showing ~15 bp DNA peeled from the histone octamer, exposing H3 N-terminus—mimicking eukaryotic nucleosome unwrapping. | Cas9-sgRNA ribonucleoprotein targets linker DNA (PAM1/PAM28) and entry-exit regions (SHL6) of nucleosomes, avoiding tightly wrapped core DNA (SHL0-5), as revealed by native-PAGE on Widom 601 nucleosomes. The cryo-EM structure (PDB: 8YNY, 4.52 Å, EMDB: EMD-39431) captures the post-cleavage ternary complex at PAM1, showing ~15 bp DNA peeled from the histone octamer, exposing H3 N-terminus—mimicking eukaryotic nucleosome unwrapping. | ||
| | ||
| - | + | ''' | |
| | ||
| - | + | Key Interactions''' | |
| - | ''' | + | |
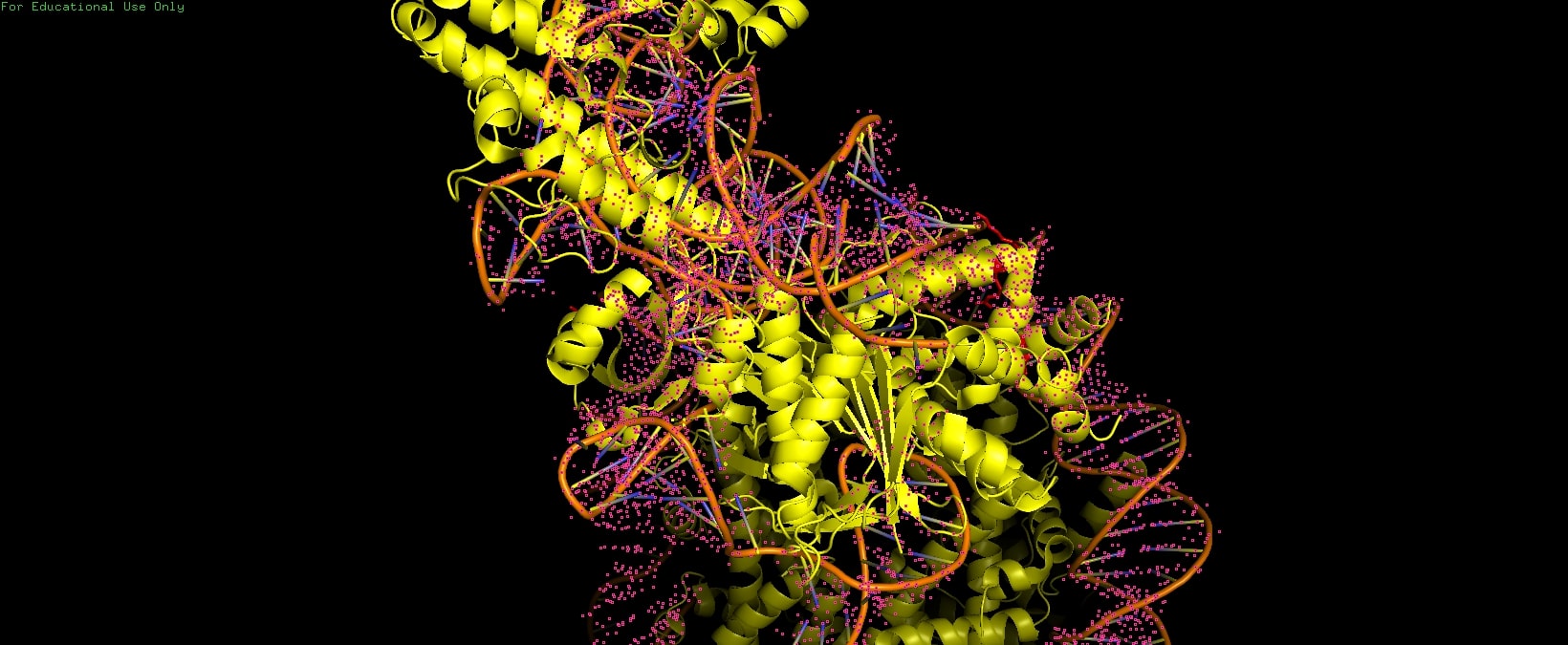
Cas9's PI domain (residues ~1100-1368) mediates multiple contacts: electrostatic histone tail interactions (non-essential for binding), PI edge lysine K1155 stabilizing the post-cleavage complex, and core DNA loops (H1264, R1298, K1300) causing inhibitory non-specific binding. Mutants disrupting PI-core DNA contacts (H1264A/R1298Q) enhance both in vitro cleavage efficiency and rice genome editing. | Cas9's PI domain (residues ~1100-1368) mediates multiple contacts: electrostatic histone tail interactions (non-essential for binding), PI edge lysine K1155 stabilizing the post-cleavage complex, and core DNA loops (H1264, R1298, K1300) causing inhibitory non-specific binding. Mutants disrupting PI-core DNA contacts (H1264A/R1298Q) enhance both in vitro cleavage efficiency and rice genome editing. | ||
| | ||
| - | | + | ''' |
| - | '''Biological Insights''' | + | Biological Insights''' |
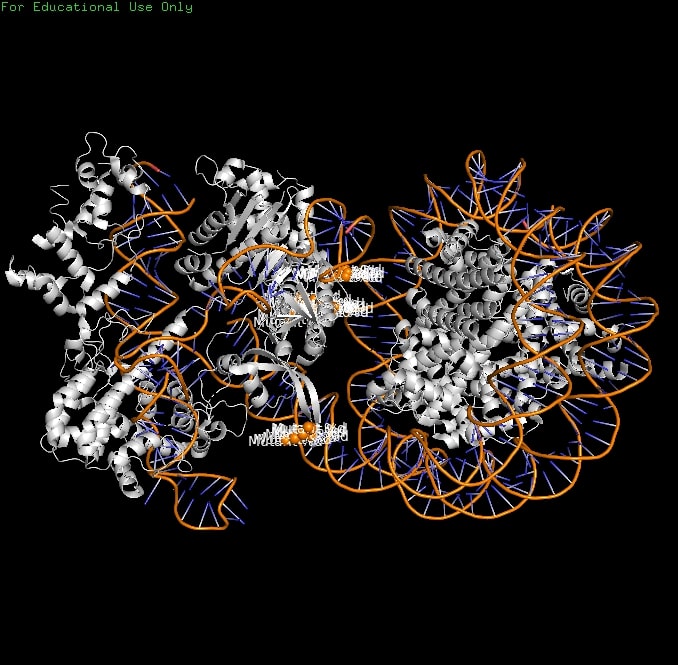
Nucleosomes inhibit Cas9 via two mechanisms: (1) DNA end inflexibility blocks access; (2) PI domain trapping restricts domain motions needed for cleavage. Entry-exit asymmetry arises from Widom601 sequence-dependent flexibility, explaining variable editing efficiency across chromatin contexts. These findings reveal Cas9's eukaryotic adaptation and guide chromatin-optimized variants for improved genome editing. | Nucleosomes inhibit Cas9 via two mechanisms: (1) DNA end inflexibility blocks access; (2) PI domain trapping restricts domain motions needed for cleavage. Entry-exit asymmetry arises from Widom601 sequence-dependent flexibility, explaining variable editing efficiency across chromatin contexts. These findings reveal Cas9's eukaryotic adaptation and guide chromatin-optimized variants for improved genome editing. | ||
| Line 19: | Line 18: | ||
''Scene 1: Overall complex'' | ''Scene 1: Overall complex'' | ||
[[Image:8YNYOverall.jpg]] | [[Image:8YNYOverall.jpg]] | ||
| + | |||
| + | ''Scene 2: PI domain contacts'' | ||
| + | [[Image:8YNYPI.jpg]] | ||
| + | '' | ||
| + | Scene 3: Mutant sites'' | ||
| + | [[Image:8YNYMutant.jpg]] | ||
Revision as of 17:48, 30 November 2025
Cas9-Nucleosome Complex (PDB: 8YNY)
Cas9-sgRNA ribonucleoprotein targets linker DNA (PAM1/PAM28) and entry-exit regions (SHL6) of nucleosomes, avoiding tightly wrapped core DNA (SHL0-5), as revealed by native-PAGE on Widom 601 nucleosomes. The cryo-EM structure (PDB: 8YNY, 4.52 Å, EMDB: EMD-39431) captures the post-cleavage ternary complex at PAM1, showing ~15 bp DNA peeled from the histone octamer, exposing H3 N-terminus—mimicking eukaryotic nucleosome unwrapping. Key Interactions
Cas9's PI domain (residues ~1100-1368) mediates multiple contacts: electrostatic histone tail interactions (non-essential for binding), PI edge lysine K1155 stabilizing the post-cleavage complex, and core DNA loops (H1264, R1298, K1300) causing inhibitory non-specific binding. Mutants disrupting PI-core DNA contacts (H1264A/R1298Q) enhance both in vitro cleavage efficiency and rice genome editing.
Biological Insights
Nucleosomes inhibit Cas9 via two mechanisms: (1) DNA end inflexibility blocks access; (2) PI domain trapping restricts domain motions needed for cleavage. Entry-exit asymmetry arises from Widom601 sequence-dependent flexibility, explaining variable editing efficiency across chromatin contexts. These findings reveal Cas9's eukaryotic adaptation and guide chromatin-optimized variants for improved genome editing.