1ysa
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1ysa.jpg|left|200px]] | [[Image:1ysa.jpg|left|200px]] | ||
| - | + | <!-- | |
| - | + | The line below this paragraph, containing "STRUCTURE_1ysa", creates the "Structure Box" on the page. | |
| - | + | You may change the PDB parameter (which sets the PDB file loaded into the applet) | |
| - | + | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | |
| - | | | + | or leave the SCENE parameter empty for the default display. |
| - | | | + | --> |
| - | + | {{STRUCTURE_1ysa| PDB=1ysa | SCENE= }} | |
| - | + | ||
| - | + | ||
| - | }} | + | |
'''THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX''' | '''THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX''' | ||
| Line 29: | Line 26: | ||
[[Category: Harrison, S C.]] | [[Category: Harrison, S C.]] | ||
[[Category: Struhl, K.]] | [[Category: Struhl, K.]] | ||
| - | [[Category: | + | [[Category: Double helix]] |
| - | [[Category: | + | [[Category: Protein-dna complex]] |
| - | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sat May 3 16:43:06 2008'' | |
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | |
Revision as of 13:43, 3 May 2008
| |||||||||
| 1ysa, resolution 2.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX
Overview
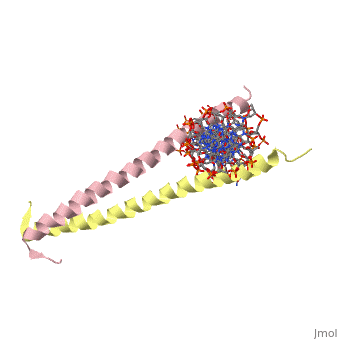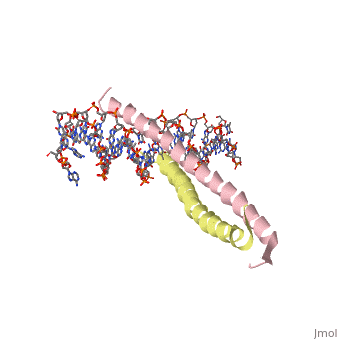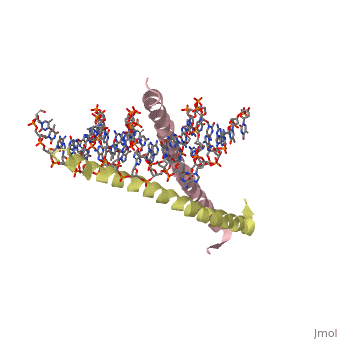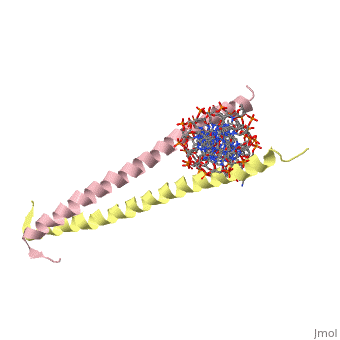
The yeast transcriptional activator GCN4 is 1 of over 30 identified eukaryotic proteins containing the basic region leucine zipper (bZIP) DNA-binding motif. We have determined the crystal structure of the GCN4 bZIP element complexed with DNA at 2.9 A resolution. The bZIP dimer is a pair of continuous alpha helices that form a parallel coiled coil over their carboxy-terminal 30 residues and gradually diverge toward their amino termini to pass through the major groove of the DNA-binding site. The coiled-coil dimerization interface is oriented almost perpendicular to the DNA axis, giving the complex the appearance of the letter T. There are no kinks or sharp bends in either bZIP monomer. Numerous contacts to DNA bases and phosphate oxygens are made by basic region residues that are conserved in the bZIP protein family. The details of the bZIP dimer interaction with DNA can explain recognition of the AP-1 site by the GCN4 protein.
About this Structure
1YSA is a Single protein structure of sequence from Saccharomyces cerevisiae. Full crystallographic information is available from OCA.
Reference
The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex., Ellenberger TE, Brandl CJ, Struhl K, Harrison SC, Cell. 1992 Dec 24;71(7):1223-37. PMID:1473154 Page seeded by OCA on Sat May 3 16:43:06 2008