3cln
From Proteopedia
| Line 1: | Line 1: | ||
| + | {{Seed}} | ||
[[Image:3cln.png|left|200px]] | [[Image:3cln.png|left|200px]] | ||
| Line 9: | Line 10: | ||
{{STRUCTURE_3cln| PDB=3cln | SCENE= }} | {{STRUCTURE_3cln| PDB=3cln | SCENE= }} | ||
| - | + | ===STRUCTURE OF CALMODULIN REFINED AT 2.2 ANGSTROMS RESOLUTION=== | |
| - | {{ | + | <!-- |
| + | The line below this paragraph, {{ABSTRACT_PUBMED_3145979}}, adds the Publication Abstract to the page | ||
| + | (as it appears on PubMed at http://www.pubmed.gov), where 3145979 is the PubMed ID number. | ||
| + | --> | ||
| + | {{ABSTRACT_PUBMED_3145979}} | ||
==About this Structure== | ==About this Structure== | ||
| - | 3CLN is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Rattus_rattus Rattus rattus]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1cln 1cln]. | + | 3CLN is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Rattus_rattus Rattus rattus]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1cln 1cln]. Additional information on 3CLN is available in a page on [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb44_1.html Calmodulin] at the RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3CLN OCA]. |
| + | |||
| + | ==Reference== | ||
| + | Structure of calmodulin refined at 2.2 A resolution., Babu YS, Bugg CE, Cook WJ, J Mol Biol. 1988 Nov 5;204(1):191-204. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/3145979 3145979] | ||
[[Category: Calmodulin]] | [[Category: Calmodulin]] | ||
[[Category: Rattus rattus]] | [[Category: Rattus rattus]] | ||
| Line 24: | Line 32: | ||
[[Category: Calcium binding protein]] | [[Category: Calcium binding protein]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Wed Jul 23 11:12:53 2008'' |
Revision as of 08:12, 23 July 2008
| |||||||||
| 3cln, resolution 2.20Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
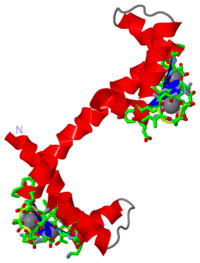
STRUCTURE OF CALMODULIN REFINED AT 2.2 ANGSTROMS RESOLUTION
The crystal structure of mammalian calmodulin has been refined at 2.2 A (1 A = 0.1 nm) resolution using a restrained least-squares method. The final crystallographic R-factor, based on 6685 reflections in the range 2.2 A less than or equal to d less than or equal to 5.0 A with intensities exceeding 2.5 sigma, is 0.175. Bond lengths and bond angles in the molecule have root-mean-square deviations from ideal values of 0.016 A and 1.7 degrees, respectively. The refined model includes residues 5 to 147, four Ca2+ and 69 water molecules per molecule of calmodulin. The electron density for residues 1 to 4 and 148 is poorly defined, and they are not included in the model. The molecule is shaped somewhat like a dumbbell, with an overall length of 65 A; the two lobes are connected by a seven-turn alpha-helix. Prominent secondary structural features include seven alpha-helices, four Ca2+-binding loops, and two short, double-stranded antiparallel beta-sheets between pairs of adjacent Ca2+-binding loops. The four Ca2+-binding domains in calmodulin have a typical EF hand conformation (helix-loop-helix) and are similar to those described in other Ca2+-binding proteins. The X-ray structure determination of calmodulin shows a large hydrophobic cleft in each half of the molecule. These hydrophobic regions probably represent the sites of interaction with many of the pharmacological agents known to bind to calmodulin.
Structure of calmodulin refined at 2.2 A resolution., Babu YS, Bugg CE, Cook WJ, J Mol Biol. 1988 Nov 5;204(1):191-204. PMID:3145979
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this Structure
3CLN is a Single protein structure of sequence from Rattus rattus. This structure supersedes the now removed PDB entry 1cln. Additional information on 3CLN is available in a page on Calmodulin at the RCSB PDB Molecule of the Month. Full crystallographic information is available from OCA.
Reference
Structure of calmodulin refined at 2.2 A resolution., Babu YS, Bugg CE, Cook WJ, J Mol Biol. 1988 Nov 5;204(1):191-204. PMID:3145979
Page seeded by OCA on Wed Jul 23 11:12:53 2008