DNA-binding protein VirE2 from Agrobacterium tumefaciens complexed with chaperone VirE1
From Proteopedia
| Line 3: | Line 3: | ||
{{STRUCTURE_3btp| PDB=3btp | SCENE=VirE1/VirE2/Vire1_vire2/1 |CAPTION=VirE1/VirE2 }} | {{STRUCTURE_3btp| PDB=3btp | SCENE=VirE1/VirE2/Vire1_vire2/1 |CAPTION=VirE1/VirE2 }} | ||
| - | + | ===Crystal structure of Agrobacterium tumefaciens VirE2 in complex with its chaperone VirE1: a novel fold and implications for DNA binding=== | |
| + | |||
<!-- | <!-- | ||
The line below this paragraph, {{ABSTRACT_PUBMED_18678909}}, adds the Publication Abstract to the page | The line below this paragraph, {{ABSTRACT_PUBMED_18678909}}, adds the Publication Abstract to the page | ||
Revision as of 11:37, 22 August 2008
Crystal structure of Agrobacterium tumefaciens VirE2 in complex with its chaperone VirE1: a novel fold and implications for DNA binding
Agrobacterium tumefaciens infects its plant hosts by a mechanism of horizontal gene transfer. This capability has led to its widespread use in artificial genetic transformation. In addition to DNA, the bacterium delivers an abundant ssDNA binding protein, VirE2, whose roles in the host include protection from cytoplasmic nucleases and adaptation for nuclear import. In Agrobacterium, VirE2 is bound to its acidic chaperone VirE1. When expressed in vitro in the absence of VirE1, VirE2 is prone to oligomerization and forms disordered filamentous aggregates. These filaments adopt an ordered solenoidal form in the presence of ssDNA, which was characterized previously by electron microscopy and three-dimensional image processing. VirE2 coexpressed in vitro with VirE1 forms a soluble heterodimer. VirE1 thus prevents VirE2 oligomerization and competes with its binding to ssDNA. We present here a crystal structure of VirE2 in complex with VirE1, showing that VirE2 is composed of two independent domains presenting a novel fold, joined by a flexible linker. Electrostatic interactions with VirE1 cement the two domains of VirE2 into a locked form. Comparison with the electron microscopy structure indicates that the VirE2 domains adopt different relative orientations. We suggest that the flexible linker between the domains enables VirE2 to accommodate its different binding partners.
Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners., Dym O, Albeck S, Unger T, Jacobovitch J, Branzburg A, Michael Y, Frenkiel-Krispin D, Wolf SG, Elbaum M, Proc Natl Acad Sci U S A. 2008 Aug 12;105(32):11170-5. Epub 2008 Aug 4. PMID:18678909
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
|
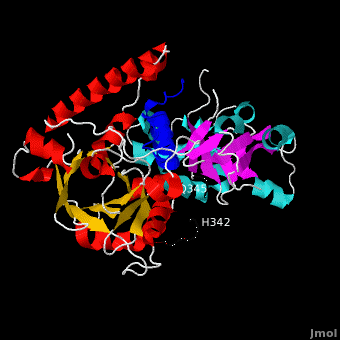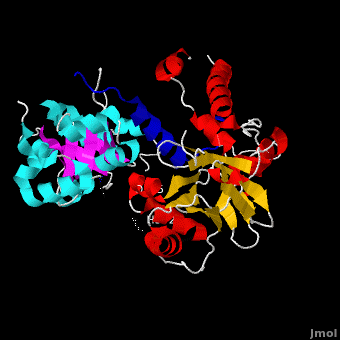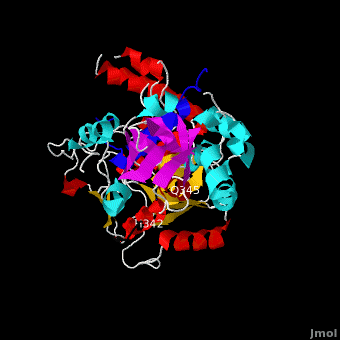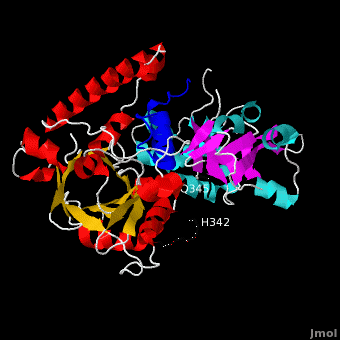
Ribbon representation of complex. The is shown in blue. The of VirE2 is depicted in red for and yellow for , while the is shown in cyan for and magenta for . The inter-domain linker (residues 337-346) is shown as a thick black line for which no electron density was observed between residues H342 and Q345. Both domains form a , composed of α-helices and β-strands resulting in a donut shape with interior and exterior . The barrel is characterized by an 8-stranded β-sheet that is closed with shear number S=8, and has a unique topology. Instead of eight repeats of the as in a (e.g. 8tim), four are present in the C-terminal domain and variations of this motif in the N-terminal domain. The topology of the VirE2 barrel in fact resembles that of the dimeric enhancer of rudimentary gene (ERH domain PDB code 1w9g). Taken together, the novel combination of architecture and topology of the VirE2 domain allows its classification as a novel fold, denoted here as the (4 ββαα motifs are shown in red, green, yellow, and magenta, respectively).
|
between VirE1 (blue) and VirE2. Residues from both domains of VirE2 form electrostatic interactions (dashed lines) with acidic residues from VirE1. N terminal domain is colored green and C terminal domain - magenta.
Alignment of the crystal structure of VirE1-VirE2 complex with the VirE2 envelope obtained by electron microscopy (EM) in the presence of ssDNA. The crystal structure of VirE2, treated as a single rigid body, was manually aligned into the envelope of one repeat of the VirE2 solenoid as determined by EM (beige). VirE1 (blue) is aligned with the solenoid axis, facing the solenoid interior (black arrow). The view is down the solenoid axis. Rotation of view by 90º perpendicular to the solenoid axis (red line), showing one VirE2 repeat viewed from the exterior of the solenoid. It is clear that while one domain of VirE2 neatly fits the envelope, the other protrudes from it (the inter-domain flexible linker is shown in black).
Schematic representation of VirE2 showing how its inter-domain flexible linker allows structural rearrangements with its different partners. The two domains of VirE2 are shown in purple and cyan linked by their inter-domain flexible linker shown in orange (A) In the presence of VirE1 (red), the two VirE2 domains are “locked” by their interaction with VirE1 fixing the inter domain linker. (B) In the absence of VirE1, the domains of VirE2 are “unlocked” and utilize the flexible inter-domain linker to rotate. (C) In the “unlocked” form, VirE2 has a strong tendency to self assemble forming N to C interactions. Due to the flexible linker, the two domains of VirE2 can adopt a range of orientations resulting in irregular filaments. (D) Upon addition of ssDNA to the filaments in C, an ordered solenoid assembly is formed (gray envelope). The ssDNA limits the degree of freedom in the linker, thereby imposing a favorable VirE2-VirE2 arrangement.
Reference
Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners., Dym O, Albeck S, Unger T, Jacobovitch J, Branzburg A, Michael Y, Frenkiel-Krispin D, Grayer Wolf S, Elbaum M., Proc Natl Acad Sci U S A. 2008 Aug 4. PMID:18678909
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Michal Harel, Eran Hodis, Tamar Unger, Jaime Prilusky, David Canner, Joel L. Sussman, Eric Martz