User:Tom Gluick/glutamine synthetase
From Proteopedia
| Line 7: | Line 7: | ||
{{STRUCTURE_2qc8 | PDB=2qc8 | SCENE= }} | {{STRUCTURE_2qc8 | PDB=2qc8 | SCENE= }} | ||
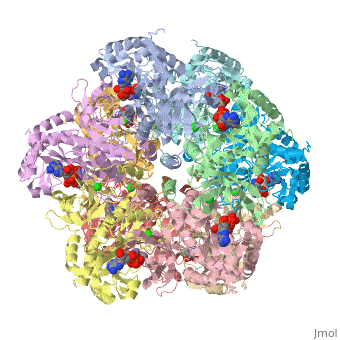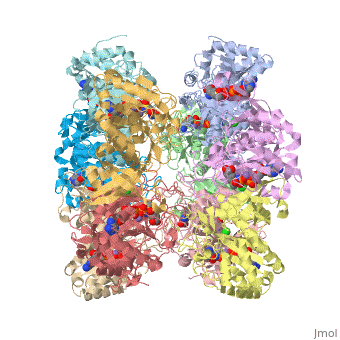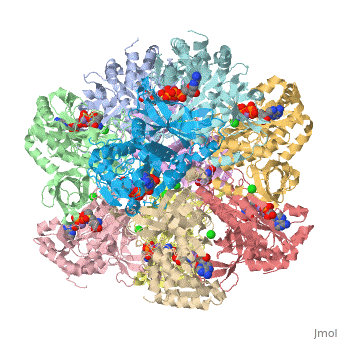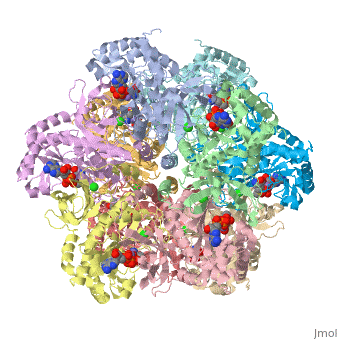
| + | The first view that is shown in the JMOL window is cartoon version of the human enzyme. The enzyme synthesizes glutamine from glutamate and ammonium ion via glutamyl-P intermediate. Details of the reaction can be viewed via clicking the PDBsum link shown in green under XXXX. Scrolling down the PDBsum page will show the reaction catalyzed by this enzyme. | ||
<scene name='User:Tom_Gluick/glutamine_synthetase/Spacefill/1'>SpaceFill version</scene> | <scene name='User:Tom_Gluick/glutamine_synthetase/Spacefill/1'>SpaceFill version</scene> | ||
Revision as of 20:28, 23 August 2008
Glutamine Synthetaser
I am trying to learn how to use this for myself and for others "Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id after the STRUCTURE_ and after PDB= to load and display another structure.
| |||||||||
| 2qc8, resolution 2.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , | ||||||||
| Gene: | GLUL, GLNS (Homo sapiens) | ||||||||
| Activity: | Glutamate--ammonia ligase, with EC number 6.3.1.2 | ||||||||
| Related: | 2ojw | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The first view that is shown in the JMOL window is cartoon version of the human enzyme. The enzyme synthesizes glutamine from glutamate and ammonium ion via glutamyl-P intermediate. Details of the reaction can be viewed via clicking the PDBsum link shown in green under XXXX. Scrolling down the PDBsum page will show the reaction catalyzed by this enzyme.
To make the scene that I show in the next link, I used the following commands in the JMOL console. The Console is accessed through the JMOL at the bottom of the figure in Show XXXX. Click on JMOL to access the pulldown menu and drag cursor to Console. The console is a place where explicit commands using the JMOL language can be used. The language takes some time to learn. However, to only show two subunit of the ten, we do type this command restrict :A, :E. This command hides the remaining 8 chains and only shows the two contiguous chains A and E. At the interface of A and E, the catalytic site resides.