This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Tom Gluick/glutamine synthetase
From Proteopedia
m (→Section 1: Using Scene Authoring Tools) |
m (→Section 1: Using Scene Authoring Tools) |
||
| Line 10: | Line 10: | ||
I have found that the tools are great for making some simple images, but I have not mastered how to make what I would call a complex image. To make a complex image I will teach you how to write your own JMOL script using the Console found in JMOL. | I have found that the tools are great for making some simple images, but I have not mastered how to make what I would call a complex image. To make a complex image I will teach you how to write your own JMOL script using the Console found in JMOL. | ||
| - | Using the Scene Authoring Tools. Below are the sequence of | + | Using the Scene Authoring Tools. Below are the sequence of commands I used to construct the following scene that is accessible in this link: <scene name='User:Tom_Gluick/glutamine_synthetase/Backbone_trace/1'>Example: Backbone Trace with ligands</scene> <br/> <br/> |
1. Open Scene authoring tools ( scroll down) <br/> | 1. Open Scene authoring tools ( scroll down) <br/> | ||
Revision as of 04:04, 25 August 2008
Glutamine Syntheteaser
Introduction
The following is a series of instructions designed to help students to complete an assignment in Biol 430 (Biological Chemistry) at the University of Maryland, Baltimore County. The instructions are divided into two sections. In the first section, images are constructed using the Scene Authoring Tools. In the second section, images are constructed using the JMOL command language. Each section is divided into three parts. In the first part: the commands are listed in order and a link is provided showing the image that is formed using the commands. In the second part, the commands are listed and the final product is shown as a still image. Your job is to use the commands to provide the final image in the Sandbox. The third part is a link showing an image where you ought to be able to construct.
| |||||||||
| 2qc8, resolution 2.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , | ||||||||
| Gene: | GLUL, GLNS (Homo sapiens) | ||||||||
| Activity: | Glutamate--ammonia ligase, with EC number 6.3.1.2 | ||||||||
| Related: | 2ojw | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
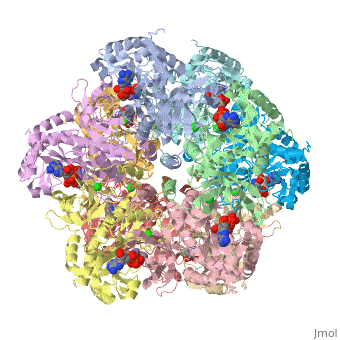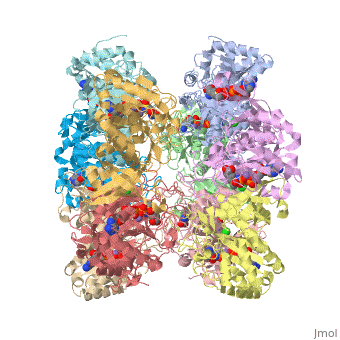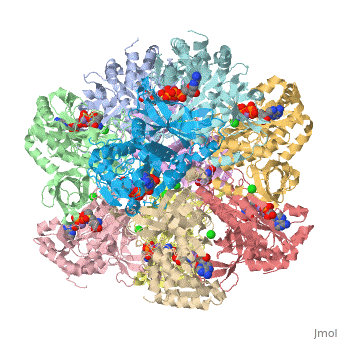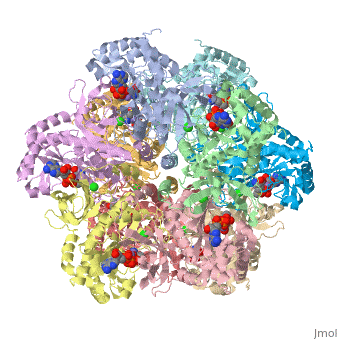
The first view that is shown in the JMOL window is cartoon version of the human enzyme. The video tutorial in [1] will give you the tools to get started and give you a great overview. The enzyme synthesizes glutamine from glutamate and ammonium ion via glutamyl-P intermediate. Details of the reaction can be viewed via clicking the PDBsum link shown in green under Resources. Scrolling down the PDBsum page will show the reaction catalyzed by this enzyme. Let's have some fun before we get into it. Proteopedia has it set up to visualize the ligands. Click on ADP or any other ligand in the green region will show cause the protein to become transparent revealing the buried ligand. Clicking on green link initial scene will return the image to the original scene.
Section 1: Using Scene Authoring Tools
I have found that the tools are great for making some simple images, but I have not mastered how to make what I would call a complex image. To make a complex image I will teach you how to write your own JMOL script using the Console found in JMOL.
Using the Scene Authoring Tools. Below are the sequence of commands I used to construct the following scene that is accessible in this link:
1. Open Scene authoring tools ( scroll down)
2. Click on the Load Molecule Tab
3. Type in 2qc8
4. Click on Selections Tab
5. Click on Select None ( this sets it up so you want to add to a selection) Select all, you use this to remove stuff from selection.
6. Highlight all protein and then click on Add to Selection
7. Click representations tab ( you can fool around here to discover the many different ways of representing a molecule) I chose Backbone and clicked on the set representation button. The ligands will be plainly visible as spacefilling models.
8. Click on colors Tab.
9. Again we are faced with many choices and you are welcome to fool around. I chose N to C rainbow. This colors the N-terminus blue and the C-terminus red, the opposite of ROY G BIV.
10. What we have done is created a script of commands to yield the representation of the molecule seen in the JMOL window. To save the script, we click on the save scene tab.
11. Now copy the Wiki script, paste it in document, put a title in the script, and insert in the document.
12. Hide the tools and hit preview to test it out.
If you like the scene, scroll down and click on Save Page; if not, then hit the back button of your browser and edit.
Now I will give you a set of instructions to follow and the final image which is shown below.
1. Return to scene authoring tools. </br>
2. Load scene-TAB and choose the Back_trace, version 1.
3. Selections/select all/halos this means hit selection tab, select all button, and check halos.
4. Representations/cartoon/set/remove halo.
5. Colors/secondary structure/background/black since the coil regions are in white.
6. Selections/select none/ligands/add to selection/halos to show
7. Representation/spacefill/remove halos
8. colors/CPK
9. Selections/select none/Mn in last box/add to selecton.
10. Colors/ochre the CPK color of Mn is difficult to discern
11. selections/select none/all protein/add to selection.
12. colors/90% transparency
13. JMOL/zoom 800%
14. save scene/copy wiki text/paste it in/name it/ preview it/save it.
The image ought to look close to this--you can see the ADP, P3S, the chloride in green, and Mn in ochre.
Image:Transpareny zoom 800.jpg
To make the scene that I show in the next link, I used the following commands in the JMOL console. The Console is accessed through the JMOL at the bottom of the figure in Show XXXX. Click on JMOL to access the pulldown menu and drag cursor to Console. The console is a place where explicit commands using the JMOL language can be used. The language takes some time to learn but it offers a lot of control. The scene authoring tools that proteopedia uses are useful for less complicated edits or when the PDB file is annotated very well with many details, like the one shown in the example movies.
To imitate what was done in this link, the following commands in the console is used use
load /cgi-bin/getpdbz?2qc8 (load molecule in scene authoring tools)
script /wiki/scripts/initialview02.spt (this is the script that provides the initial view)
spin off (spin is set to off by default)
select protein (command typed in to select all residues called protein by the PDB file)
color green ( colors the selected protein green)
color green translucent (200) (this sets the translucency to 200)
restrict protein, ADP ( restrict only selects and views those selected, not syntax, all none selected residues are not shown)
zoom 150 (this zooms the image by 150%)
spin (sets the image spinning)
Next, I went back to scene authoring tools, and clicked on labels. I filled in the caption to label the figure with ADP. Remember to click set label!
Now, you give this script a whirl. In this case, the protein is red( bad color as you will see), its translucency is set to 200. Restrict to protein and P3S, and zoom 200; setting the spin is optional. The scene as a still ought to look something like this image: Image:Translucent red P3S.jpg
However, to only show two subunit of the ten, we do type this command restrict :A, :E. This command hides the remaining 8 chains and only shows the two contiguous chains A and E. At the interface of A and E, the catalytic site resides.