User:Tom Gluick/glutamine synthetase
From Proteopedia
(Undo revision 760116 by Tom Gluick (Talk)) |
(→'''Exercise 1: USING SCENE AUTHORING TOOLS WITH EXPLANATIONS''') |
||
| Line 15: | Line 15: | ||
=== '''Exercise 1: USING SCENE AUTHORING TOOLS WITH EXPLANATIONS''' === | === '''Exercise 1: USING SCENE AUTHORING TOOLS WITH EXPLANATIONS''' === | ||
| - | Below are the sequence of commands I used to construct | + | Below are the sequence of commands I used to construct a scene that is accessible in the green link below the script. The image portrays the enzyme in differently than before. Here, the protein is reduced to a tracing of the protein backbone which reveals the ligands, ADP, P3S, Cl<sup>-</sup> and Mn<sup>2+</sup> as spacefill models within the protein.<br/> <applet load='2qc8' size='300' frame='true' align='right' caption='Practice Applet' /><br/><br/> |
1. '''Open Scene authoring tools''' ( scroll down-this allows you to load an image from the PDB or a previously saved scene). <br/> | 1. '''Open Scene authoring tools''' ( scroll down-this allows you to load an image from the PDB or a previously saved scene). <br/> | ||
| Line 26: | Line 26: | ||
8. '''Click on colors Tab.''' <br/> | 8. '''Click on colors Tab.''' <br/> | ||
9. Again we are faced with many choices and you are welcome to fool around. I chose N to C rainbow. This colors the N-terminus blue and the C-terminus red, the opposite of ROY G BIV. <br/> | 9. Again we are faced with many choices and you are welcome to fool around. I chose N to C rainbow. This colors the N-terminus blue and the C-terminus red, the opposite of ROY G BIV. <br/> | ||
| - | 10. What we have done is created a script of commands to yield the representation of the molecule seen in the JMOL window. '''To save the script, we click on the save scene tab'''.<br/> | + | 10. What we have done is created a script of commands to yield the representation of the molecule seen in the JMOL window. '''To save the script, we click on the save scene tab'''. Name and describe the script in the appropriate boxes.<br/> |
| - | 11. Now copy the Wiki script, paste it in document, put a title in the script, and insert in the document.<br/> | + | 11. Now copy the Wiki script, paste it in document, put a title in the script, and insert in the document. The wiki script has this format: <nowiki><scene name='User:Tom_Gluick/glutamine_synthetase/Backbone_trace/1'>Insert text here</scene></nowiki> I inserted the text: Example: Backbone Trace with ligand. <br/> |
12. Hide the tools and hit preview to test it out.<br/> | 12. Hide the tools and hit preview to test it out.<br/> | ||
When scene is to your liking, scroll down and click on Save Page; if not, then hit the back button of your browser and edit. <br/> | When scene is to your liking, scroll down and click on Save Page; if not, then hit the back button of your browser and edit. <br/> | ||
| + | The wiki script called <scene name='User:Tom_Gluick/glutamine_synthetase/Backbone_trace/1'>Example: Backbone Trace with ligands</scene> was inserted within this sentence. | ||
=== '''Exercise 2: LIGAND SELECTION AND REPRESENTATION''' === | === '''Exercise 2: LIGAND SELECTION AND REPRESENTATION''' === | ||
Revision as of 17:12, 14 September 2008
Contents |
Glutamine Syntheteaser
| |||||||||
| 2qc8, resolution 2.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , | ||||||||
| Gene: | GLUL, GLNS (Homo sapiens) | ||||||||
| Activity: | Glutamate--ammonia ligase, with EC number 6.3.1.2 | ||||||||
| Related: | 2ojw | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Glutamine synthetase is an key enzyme in amino acid metabolism found in all the phyla. The cartoon shown in the JMOL applet on the right hand side of the page is an image of the human enzyme. More information is found in the human glutamine synthetase stub[1]. Both the human and the plant enzyme are a dimer of stacked pentamers[2]The crystal structure of the Salmonella typhimurium enzyme has been solved and published in 1989, and its regulation is a subject of textbook discourse[3]. Three different classes of GS are classified based on oligomeric structure[4]. This information can be also accessed by clicking the PFAM links in structural annotation resources..
The enzyme synthesizes glutamine from glutamate and ammonium ion via glutamyl-P intermediate. In humans, defects in GLUL ( the gene encoding for gluatmine synthetase) is found in a rare congenital disorder leading to brain malformation and death in neonates[5]. Details of the reaction and the enzyme can be obtained through clicking the PDBsum link shown in green under Resources and exploring the links, especially Brenda. Scrolling down the PDBsum page will show the reaction catalyzed by this enzyme.
Before you start the project, I suggest you view the video tutorial in [6] to provide you with the tools to get started and give you a great overview of the power of proteopdia. Let's have some fun before we get into it. Proteopedia has it set up to visualize the ligands. Click on ADP or any other ligand in the green region will show cause the protein to become transparent revealing the buried ligand. Clicking on green link initial scene will return the image to the original scene.
Introduction to the Project
The following is a series of instructions designed to help students to complete an assignment in Biol 430 (Biological Chemistry) at the University of Maryland, Baltimore County. The instructions are divided into two sections. In the first section, images are constructed using the Scene Authoring Tools. In the second section, images are constructed using the JMOL command language. Each section is divided into three parts. In the first part: the commands are listed in order and a link is provided showing the image that is formed using the commands. In the second part, the commands are listed and the final product is shown as a still image. Your job is to use the commands to provide the final image in the Sandbox. The third part is a link showing an image where you ought to be able to construct.
Section 1: Using Scene Authoring Tools
I have found that the tools are great for making some simple images, but I have not mastered how to make what I would call a complex image. To make a complex image I will teach you how to write your own JMOL script using the Console found in JMOL.
Exercise 1: USING SCENE AUTHORING TOOLS WITH EXPLANATIONS
Below are the sequence of commands I used to construct a scene that is accessible in the green link below the script. The image portrays the enzyme in differently than before. Here, the protein is reduced to a tracing of the protein backbone which reveals the ligands, ADP, P3S, Cl- and Mn2+ as spacefill models within the protein.
|
1. Open Scene authoring tools ( scroll down-this allows you to load an image from the PDB or a previously saved scene).
2. Click on the Load Molecule Tab (one of the options. This one allows you to load the PDB file).
3. Type in 2qc8(this XXXXX signifies the file containing the coordinates to describe a crystal structure of human glutamine synthetase.).
4. Click on Selections Tab (this tab will bring up a new window that will let you select all, none or parts of the structure).
5. Click on Select None ( this sets it up so you want to add to a selection) Select all, you use this to remove stuff from selection.
6. Highlight the selection in the large box called: all protein, then click on Add to Selection.(this will add the protein, and hopefully nothing else to be subject to the next set of operations).
7. Click representations tab ( you can fool around here to discover the many different ways of representing a molecule) I chose Backbone and clicked on the set representation button. The ligands will be plainly visible as spacefilling models.
8. Click on colors Tab.
9. Again we are faced with many choices and you are welcome to fool around. I chose N to C rainbow. This colors the N-terminus blue and the C-terminus red, the opposite of ROY G BIV.
10. What we have done is created a script of commands to yield the representation of the molecule seen in the JMOL window. To save the script, we click on the save scene tab. Name and describe the script in the appropriate boxes.
11. Now copy the Wiki script, paste it in document, put a title in the script, and insert in the document. The wiki script has this format: <scene name='User:Tom_Gluick/glutamine_synthetase/Backbone_trace/1'>Insert text here</scene> I inserted the text: Example: Backbone Trace with ligand.
12. Hide the tools and hit preview to test it out.
When scene is to your liking, scroll down and click on Save Page; if not, then hit the back button of your browser and edit.
The wiki script called was inserted within this sentence.
Exercise 2: LIGAND SELECTION AND REPRESENTATION
Now the following set of commands will provide an image that highlights the ligands and ions found in the crystal structure in a nearly transparent protein. In this assignment,your goal is to generate the image you shown below as still image.
1. Return to scene authoring tools.
2. Load scene-TAB and choose the Back_trace, version 1( of course you can load the molecule again using the PDB code).
3. Selections/select all/halos this means hit selection tab, select all button, and check halos.
4. Representations/cartoon/set/remove halo.
5. Colors/secondary structure/background/black since the coil regions are in white.
6. Selections/select none/ligands/add to selection/halos to show
7. Representation/spacefill/remove halos
8. Colors/CPK
9. Selections/select none/Mn in last box/add to selecton.
10. Colors/ochre the CPK color of Mn is difficult to discern
11. Selections/select none/all protein/add to selection.
12. Colors/90% transparency.
13. JMOL/zoom 800%.
14. Save scene/copy wiki text/paste it in/name it/ preview it/save it.
|
Image:Transpareny zoom 800.jpg
you can see the ADP, P3S, the chloride in green, and Mn in ochre.
Insert WIKI Script here:
Exercise 3: MORE SELECTIONS AND REPRESENTATIONS
Using the fields in Selection menu to pick individual amino acid residues and ligands. In this exercise, you are to experiment with using the fields in the selection menu to pinpoint individual amino acid residues and ligands. In this exercise, you will construct an image that shows all residues that are within 8 angstroms from the active site in subunit J. The amino acid residues are wireframe 100, and the ADP and P3S are spacefull. I used the following commands. I will provide a few commands for you to try and insert the WIki script if you so desired:
1. Load 2qc8
2. Selection/Select none/limit to Chain, J/limit to residue number 501,601/add/select within distance 8/go/halo. (This command selects for residue number 501, which is ADP and 601 which is P3S in subunit J
3. Invert(This selects for everything else, which we will delete using the next set of commands).
4. Representation/Hide/-Halo(Now, you should see the ADP, P3S and the amino acid residues as ribbons within 8 angstroms).
5. Selection/Invert/Center/Zoom(hold down Shift key and Drag across image)(selects for image that is shown and makes it bigger).
6. Representation/StickWireframe 100/set(these convert all to wireframe representation).
7. Selection/select none/J, 501,601/add(these select ADP and P3S only) .
8. Representation/Spacefill(the ligands are now spacefill representaton surrounded by wireframe)
Insert WIKI script here: to see this image in the applet above.
Exercise 4: GOING SOLO
Construct an image in the applet based on the following description. Show a single subunit without ligands or solvent in the Applet shown to the left. Represent the single subunit as spacefill model. The answer will be posted on-line in Blackboard.
Section 2. Using JMOL Commands in the Console
If you scroll up to the original JMOL applet, and click on ADP, you will notice that the image fades allowing the ligand to become prominent as it zooms in and out while the image spins. First I dare you to try to do this using the Screen Authoring Tools;
|
In the following paragraphs I will illustrate how to use several commands in JMOL which avoid some steps in the SAT. The commands i will illustrate are zoomto, select, restrict, color, translucent and... . make the scene using JMOL command language in the JMOL console. The Console is accessed through the JMOL at the bottom of the figure in the JMOL Console Practice Applet. Click on JMOL to access the pulldown menu and drag cursor to Console. The console is a place where explicit commands using the JMOL language can be used. The language takes some time to learn but it offers a lot of control. The scene authorOpening tools that proteopedia uses are useful for less complicated edits or when the PDB file is annotated very well with many details, like the one shown in the example movies.
Open Screen authoring tools<br/>
load molecule/by PDB code 2qc8/click load--this will post load /cgi-bin/getpdbz?2qc8 with script /wiki/scripts/initialview02.spt
AND spin off IF YOU PRESS THE HISTORY TAB IN THE CONSOLE.
click on JMOL frank to reveal a pulldown menu; choose console the console opens
select protein--using this command selects only those atoms called protein
color gray--this will color the selected residues gray; any color after color can be used.
color translucent 200--this command will make the selected residues translucent--the higher the number the more translucency
restrict protein, ADP--this is the restrict command that restricts the display--the statement says to restrict protein and ADP (note comma) meaning only protein and ADP are diaplayed
zoomto 3 150--this command can be used to zoom in or out. the first number says zoom occurs in 3 sec and the second the percentage increase or decrease, in this case 150%
set echo bottom center; font echo 20 bold; color echo black--this sets the label format to bottom center, font details and color
echo "Adenosine Diphosphate"--this is the label
Click save screen in SAT and select Spin ON
The WIKI appears and copy it and place it in the text.
Now, you ought to give this a whirl. If you are up to it, construct a script that would zoom and spin; translucency 200, color protein orange, color P3S black restrict to P3S and protein. Your target as a still figure ought to look similar to the figure shown here.
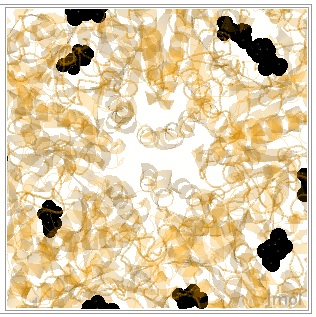
|
The commands to are listed below; these commands illustrate various ways to label amino acid residues or ligands. The script also uses the restrict and select commands, turns off spacefill and cartoon and introduces wireframe. You are also given the command to center rotation upon a single residue. You are introduced to new coloring schemes and introduced to new code to describe individual atoms of amino acids.
load /cgi-bin/getpdbz?2qc8
script /wiki/scripts/initialview02.spt
spin off
loadedfileprev = "2QC8" This command loads the molecule in SAT.
zoomto 2 800 Expands the molecule in 2 seconds 800 per cent.
select all
wireframe 0.3 Sets the width of the wire frame model
cartoon off Turns the cartoon off leaving the wireframe behind. </br>
spacefill off Turns off the spacefill function now the ligands are wireframe.
restrict 324A, 338A, 134A, 501A, 203A, 257A, 601A Restricts display to these residues only.
center 203A Centers the display on amino acid 203A
color amino Colors selected amino acid with color corresponding color.
select 501A
color cpk Colors selected amino acid with CPK colors.
select 601A
color CPK
select 401A, 402A, 403A
spacefill
spacefill 50% Decreases the size of the spacefilled molecule or atom to 50% of default size.
label SER Labels all the residues of 257A SER.
select 324A.CA Select 324 amino acid residue C alpha atom.
label ARG; color black label the above ARG and color it black.
select 338A.CA; color label black; font label 20 bold; label GLU the font label is set to 20 and is bold
select 203A.CA; color label black; font label 20 bold; label GLU
select 203A
color CPK
select 501A.O
set picking on
select 501A.N7
label ADP
select 401A, 402A, 403A
label Mn
select 601A.SD; color label black; font label 20 bold; label P3S
select GLU134A.CA; ; color label black; font label 20 bold; label GLU
spin on