This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2g2u
From Proteopedia
OCA (Talk | contribs)
(New page: 200px<br /><applet load="2g2u" size="450" color="white" frame="true" align="right" spinBox="true" caption="2g2u, resolution 1.60Å" /> '''Crystal Structure of...)
Next diff →
Revision as of 08:47, 21 November 2007
|
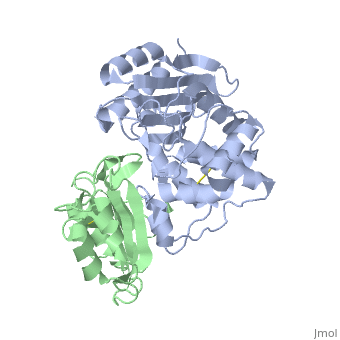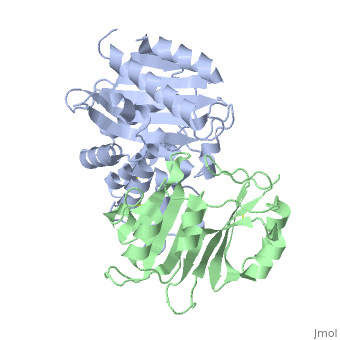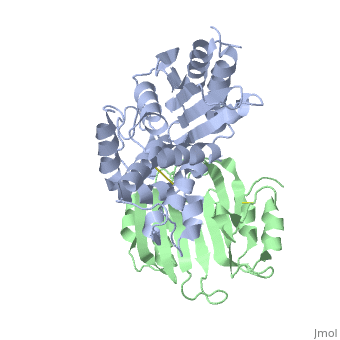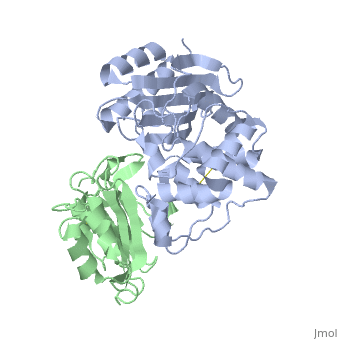
Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex
Overview
Beta-lactamase inhibitor protein (BLIP) binds a variety of class A, beta-lactamases with affinities ranging from micromolar to picomolar., Whereas the TEM-1 and SHV-1 beta-lactamases are almost structurally, identical, BLIP binds TEM-1 approximately 1000-fold tighter than SHV-1., Determining the underlying source of this affinity difference is important, for understanding the molecular basis of beta-lactamase inhibition and, mechanisms of protein-protein interface specificity and affinity. Here we, present the 1.6A resolution crystal structure of SHV-1.BLIP. In addition, a point mutation was identified, SHV D104E, that increases SHV.BLIP, binding affinity from micromolar to nanomolar. Comparison of the, SHV-1.BLIP structure with the published TEM-1.BLIP structure suggests that, the increased volume of Glu-104 stabilizes a key binding loop in the, interface. Solution of the 1.8A SHV D104K.BLIP crystal structure, identifies a novel conformation in which this binding loop is removed from, the interface. Using these structural data, we evaluated the ability of, EGAD, a program developed for computational protein design, to calculate, changes in the stability of mutant beta-lactamase.BLIP complexes. Changes, in binding affinity were calculated within an error of 1.6 kcal/mol of the, experimental values for 112 mutations at the TEM-1.BLIP interface and, within an error of 2.2 kcal/mol for 24 mutations at the SHV-1.BLIP, interface. The reasonable success of EGAD in predicting changes in, interface stability is a promising step toward understanding the stability, of the beta-lactamase.BLIP complexes and computationally assisted design, of tight binding BLIP variants.
About this Structure
2G2U is a Protein complex structure of sequences from Klebsiella pneumoniae and Streptomyces clavuligerus. Active as Beta-lactamase, with EC number 3.5.2.6 Full crystallographic information is available from OCA.
Reference
Structural and computational characterization of the SHV-1 beta-lactamase-beta-lactamase inhibitor protein interface., Reynolds KA, Thomson JM, Corbett KD, Bethel CR, Berger JM, Kirsch JF, Bonomo RA, Handel TM, J Biol Chem. 2006 Sep 8;281(36):26745-53. Epub 2006 Jun 29. PMID:16809340
Page seeded by OCA on Wed Nov 21 10:55:12 2007