User:Lois A. Fridmann/Sandbox 3
From Proteopedia
| Line 20: | Line 20: | ||
==Structural and Chemical Details of C-terminal mutant Tsg101== | ==Structural and Chemical Details of C-terminal mutant Tsg101== | ||
{{STRUCTURE_1m4p| PDB=1m4p | SCENE= }} | {{STRUCTURE_1m4p| PDB=1m4p | SCENE= }} | ||
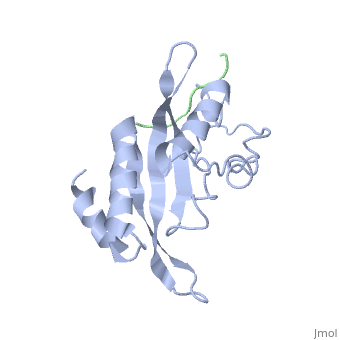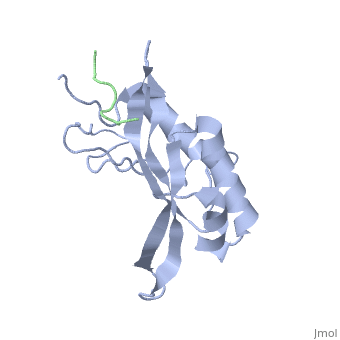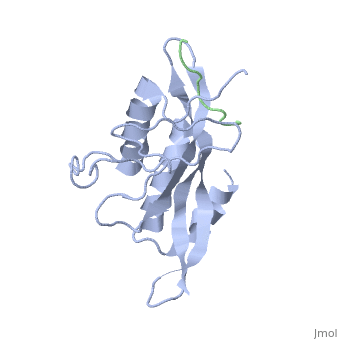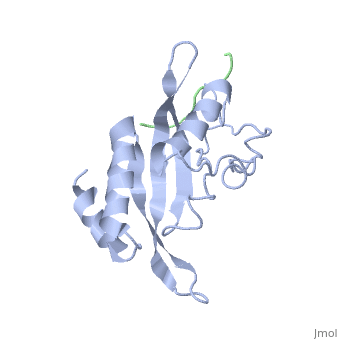
| - | The specific HIV Tsg101 Ubiquitin E2 Variant (UEV) domain model represents the wild type version of the Tsg101 UEV domain with specific highlights of the PTAP binding site featured as a green backbone, three residues in the PTAP binding site (<scene name='User:Lois_A._Fridmann/Sandbox_3/M95/2'>M95</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y63/1'>Y63</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y68/1'>Y68</scene>), which are also colored green, four C-terminal domain residues <scene name='User:Lois_A._Fridmann/Sandbox_3/Y110/1'>Y110</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y113/1'>Y113</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/K118/1'>K118</scene> colored orange and W117 colored yellow, and two residues included in the Ubiquitin binding site featured as a cyan colored backbone (V43, F88). | + | The specific HIV Tsg101 Ubiquitin E2 Variant (UEV) domain model represents the wild type version of the Tsg101 UEV domain with specific highlights of the PTAP binding site featured as a green backbone, three residues in the PTAP binding site (<scene name='User:Lois_A._Fridmann/Sandbox_3/M95/2'>M95</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y63/1'>Y63</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y68/1'>Y68</scene>), which are also colored green, four C-terminal domain residues <scene name='User:Lois_A._Fridmann/Sandbox_3/Y110/1'>Y110</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y113/1'>Y113</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/K118/1'>K118</scene> colored orange and name='User:Lois_A._Fridmann/Sandbox_3/W117/1'>W117</scene> colored yellow, and two residues included in the Ubiquitin binding site featured as a cyan colored backbone (<scene name='User:Lois_A._Fridmann/Sandbox_3/Y43/1'>V43</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/F88/1'>F88</scene>). |
The three residues featured in the PTAP binding pocket exhibit potential areas where mutations can occur that results in the elimination or reduction of the HIV-1 Gag protein binding. The two residues featured in the Ubiquitin binding pocket feature potential areas where mutations can occur that inhibits Ubiquitination of the Tsg101 or the p6 region of the Gag binding to the PTAP binding pocket. However, the main focus of the model is the C-terminal region of the UEV domain where the mutation of certain amino acid residues, which are not directly involved in the PTAP binding pocket result in the elimination or reduction of binding to the Gag protein. A mutation of W117 (yellow residues), which is conserved in all E2 enzymes, to A117 eliminates binding entirely. Mutations of Y110, Y113 and K118 (orange residues), which are unique to the Tsg101, to W110, V113 and A118 reduce binding by 31%, 33% and 50% respectively. | The three residues featured in the PTAP binding pocket exhibit potential areas where mutations can occur that results in the elimination or reduction of the HIV-1 Gag protein binding. The two residues featured in the Ubiquitin binding pocket feature potential areas where mutations can occur that inhibits Ubiquitination of the Tsg101 or the p6 region of the Gag binding to the PTAP binding pocket. However, the main focus of the model is the C-terminal region of the UEV domain where the mutation of certain amino acid residues, which are not directly involved in the PTAP binding pocket result in the elimination or reduction of binding to the Gag protein. A mutation of W117 (yellow residues), which is conserved in all E2 enzymes, to A117 eliminates binding entirely. Mutations of Y110, Y113 and K118 (orange residues), which are unique to the Tsg101, to W110, V113 and A118 reduce binding by 31%, 33% and 50% respectively. | ||
Revision as of 19:48, 13 April 2009
Tsg101 UEV Domain with a HIV-1 PTAP binding pocket - Residue Mutations in the C-Terminal End
Contents |
Background and History of HIV
The Human Immunodefiency Virus (HIV) is the causative agent of acquired immunodefiency syndrom (AIDS) in which the immune system of a human being begins to fail, leading to life threatening infections.
Tumor susceptibility gene 101, also known as Tsg101, is a human gene that encodes for a cellular protein of the same name. Tsg101 plays an important role in the pathogenesis of HIV. In infected cells, Tsg101 functions in the biogenesis of the multivesicular body (MVB), which suggests that HIV may bind Tsg101 in order to gain access to the downstream machinery that catalyzes MVB vesicle budding.
Group-specific antigen is the genetic material that codes for the core structural proteins of a retrovirus. Gag proteins are encoded by the gag gene, and provide structural elements of the virus.
The protein encoded by this gene belongs to a group of apparently inactive homologs of ubiquitin-conjugating enzymes. The gene product contains a coiled-coil domain that interacts with tathmin, a cytosolic phosphoprotein implicated in tumorigenesis. The protein may play a role in cell growth and differentiation and act as a negative growth regulator. In vitro steady-state expression of this tumor susceptibility gene appears to be important for maintenance of genomic stability and cell cycle regulation. Mutations and alternative splicing in this gene occur in high frequency in breast cancer and suggest that defects occur during breast cancer tumorigenesis and/or progression.
Tsg101 belongs to a group of apparently inactive homologs of ubiquitin-conjugating enzymes. Tsg101 contains a coiled-coil domain that interacts with stathmin, a cytosolic phosphoprotein implicated in tumorigenesis. Tsg101 may play a role in cell growth. Tsg101 is a regulator of vesicular trafficking process. Required for the sorting of endocytic ubiquitinated cargos into ultivesicular bodies. May be involved in cell growth and differentiation. It acts as a negative growth regulator involved in the budding of many viruses. The UEV domain is required for the interaction of the complex with ubiquitin. It also mediates the interaction with PTAP/PSAP motifs of HIV-1 p6 protein and human spumaretrovirus Gag protein. Tsg101 reacts with human. Ubiquitination inactivates Tsg101, possibly by regulating its shuttling between an active membrane-bound protein and an inactive soluble form. Tsg101 belongs to the ubiquitin-conjugating enzyme family. UEV subfamily.
(Generalized to other viruses: A large number of viruses in other families conserve the PTAP binding site, conserving something that is affective for other viruses)
Structural and Chemical Details of C-terminal mutant Tsg101
| |||||||||
| 1m4p, 20 NMR models () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: | tumor susceptibility gene 101 (Homo sapiens), Gag (Human immunodeficiency virus 1) | ||||||||
| Related: | 1kpp, 1kpq, 1m4q | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The specific HIV Tsg101 Ubiquitin E2 Variant (UEV) domain model represents the wild type version of the Tsg101 UEV domain with specific highlights of the PTAP binding site featured as a green backbone, three residues in the PTAP binding site (, , ), which are also colored green, four C-terminal domain residues , , colored orange and name='User:Lois_A._Fridmann/Sandbox_3/W117/1'>W117</scene> colored yellow, and two residues included in the Ubiquitin binding site featured as a cyan colored backbone (, ).
The three residues featured in the PTAP binding pocket exhibit potential areas where mutations can occur that results in the elimination or reduction of the HIV-1 Gag protein binding. The two residues featured in the Ubiquitin binding pocket feature potential areas where mutations can occur that inhibits Ubiquitination of the Tsg101 or the p6 region of the Gag binding to the PTAP binding pocket. However, the main focus of the model is the C-terminal region of the UEV domain where the mutation of certain amino acid residues, which are not directly involved in the PTAP binding pocket result in the elimination or reduction of binding to the Gag protein. A mutation of W117 (yellow residues), which is conserved in all E2 enzymes, to A117 eliminates binding entirely. Mutations of Y110, Y113 and K118 (orange residues), which are unique to the Tsg101, to W110, V113 and A118 reduce binding by 31%, 33% and 50% respectively.
Additional Mutations
Contributions to Future Research and Therapies
Identification of mutations of various residues within and surrounding the PTAP binding site that inhibit HIV-1 Gag binding can help provide important information and avenues for future research on viral treatment and vaccination.
References
1. Pornillos O, Alam S, Davis D, Sundquist W. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nature Structural Biology 9:812 – 817.
2. Tavassoli A, Lu Q, Gam J, Pan H, Benkovic S, Cohen S. 2008. Inhibition of HIV Budding by a Genetically Selected Cyclic Peptide Targeting the Gag-TSG101 Interaction. American Chemical Society, Chemical Biology 3:757 - 764.
3. VerPlank L, Bouamr F, LaGrassa T, Agresta B, Kikonyogo A, Leis J, Carter C. 2001. Tsg101, a homolgue of ubiquitin-conjugating (E2) enzymes, binds the L domain of HIV type 1 Pr55Gag. Proceeding of the National Academy of Science 98:7724 – 7729.
4.