User:Matt Whelihan
From Proteopedia
| Line 2: | Line 2: | ||
== ''' Firefly Luciferase''' == | == ''' Firefly Luciferase''' == | ||
Bioluminescence is the process by which living organisms convert chemical energy into photons of light and it is widely distributed throughout the animals, plants and fungi <ref>PMID:6358519</ref>. Species use bioluminescence as a survival tool in mating, defense and hunting. The one thing that all bioluminescent species have in common is that they all catalyze the reaction with an enzyme generically called a luciferase. All luciferases oxidize a substrate, which then decays back to the ground state while emitting a photon of light. This process is incredibly efficient with almost one photon of light produced per oxidation. While all lucifeases oxidize their substrates, the cofactors involved and reaction pathways used, vary widely <ref>PMID:775940</ref><ref>PMID:14444706</ref>. | Bioluminescence is the process by which living organisms convert chemical energy into photons of light and it is widely distributed throughout the animals, plants and fungi <ref>PMID:6358519</ref>. Species use bioluminescence as a survival tool in mating, defense and hunting. The one thing that all bioluminescent species have in common is that they all catalyze the reaction with an enzyme generically called a luciferase. All luciferases oxidize a substrate, which then decays back to the ground state while emitting a photon of light. This process is incredibly efficient with almost one photon of light produced per oxidation. While all lucifeases oxidize their substrates, the cofactors involved and reaction pathways used, vary widely <ref>PMID:775940</ref><ref>PMID:14444706</ref>. | ||
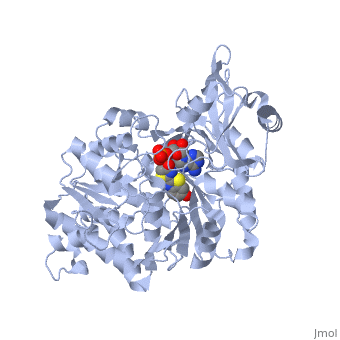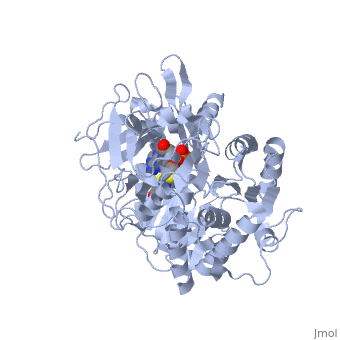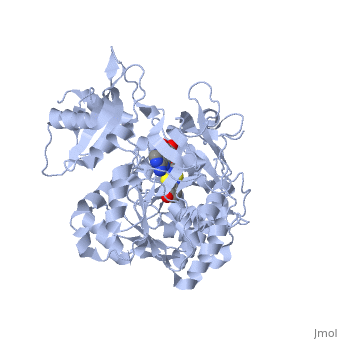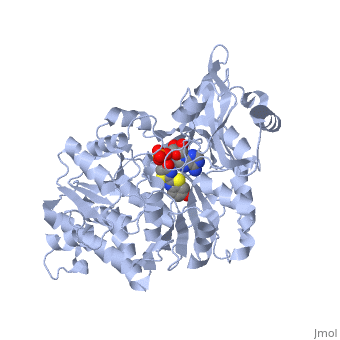
| - | + | The most studied of all luciferases is that of the North American <scene name='User:Matt_Whelihan/Rainbow_n-c/1'>Firefly</scene> Photinus Pyralis. This particular luciferase enzyme is located in the light emitting organ known as the lantern in the abdomen of the beetle. Firefly larvae glow green to ward off predators and adult fireflies use this mechanism of bioluminescence to attract mates. Luciferase binds ATP/Mg+ and D-luciferin and oxidizes it to Oxyluciferin with the products of one photon of yellow-green light, pyrophosphate, AMP and CO2<ref>PMID:775940</ref>. | |
Firefly luciferase shares significant primary sequence and mechanistic similarities with peptide synthetases and acyl-CoA ligases. These enzymes catalyze analogous activation reactions between ATP and the carboxyl moiety of their substrates <ref>PMID:1447981</ref>. An analysis of homologous sequences reveals 5 amino acids (Gly200, Lys206, Glu344, Asp422, Arg437, Gly446 | Firefly luciferase shares significant primary sequence and mechanistic similarities with peptide synthetases and acyl-CoA ligases. These enzymes catalyze analogous activation reactions between ATP and the carboxyl moiety of their substrates <ref>PMID:1447981</ref>. An analysis of homologous sequences reveals 5 amino acids (Gly200, Lys206, Glu344, Asp422, Arg437, Gly446 | ||
and Glu455)that are absolutely conserved in the sequences and these residues play a crucial role in ATP binding. | and Glu455)that are absolutely conserved in the sequences and these residues play a crucial role in ATP binding. | ||
| - | + | The major portion of the <scene name='User:Matt_Whelihan/Secondary_structure/1'>structure</scene>, comprising residues 4–436, consists of a compact domain containing a distorted antiparallel b-barrel and two b-sheets, which are flanked on either side by a-helices. The C terminus of the protein (440–544) forms a small separate a+b domain.<ref>PMID:8805533</ref> | |
<scene name='User:Matt_Whelihan/Active_site_residues/1'>active site</scene> | <scene name='User:Matt_Whelihan/Active_site_residues/1'>active site</scene> | ||
Revision as of 02:42, 20 April 2009
Firefly Luciferase
Bioluminescence is the process by which living organisms convert chemical energy into photons of light and it is widely distributed throughout the animals, plants and fungi [1]. Species use bioluminescence as a survival tool in mating, defense and hunting. The one thing that all bioluminescent species have in common is that they all catalyze the reaction with an enzyme generically called a luciferase. All luciferases oxidize a substrate, which then decays back to the ground state while emitting a photon of light. This process is incredibly efficient with almost one photon of light produced per oxidation. While all lucifeases oxidize their substrates, the cofactors involved and reaction pathways used, vary widely [2][3]. The most studied of all luciferases is that of the North American Photinus Pyralis. This particular luciferase enzyme is located in the light emitting organ known as the lantern in the abdomen of the beetle. Firefly larvae glow green to ward off predators and adult fireflies use this mechanism of bioluminescence to attract mates. Luciferase binds ATP/Mg+ and D-luciferin and oxidizes it to Oxyluciferin with the products of one photon of yellow-green light, pyrophosphate, AMP and CO2[4]. Firefly luciferase shares significant primary sequence and mechanistic similarities with peptide synthetases and acyl-CoA ligases. These enzymes catalyze analogous activation reactions between ATP and the carboxyl moiety of their substrates [5]. An analysis of homologous sequences reveals 5 amino acids (Gly200, Lys206, Glu344, Asp422, Arg437, Gly446 and Glu455)that are absolutely conserved in the sequences and these residues play a crucial role in ATP binding. The major portion of the , comprising residues 4–436, consists of a compact domain containing a distorted antiparallel b-barrel and two b-sheets, which are flanked on either side by a-helices. The C terminus of the protein (440–544) forms a small separate a+b domain.[6]
|