User:Matt Whelihan
From Proteopedia
| Line 5: | Line 5: | ||
Bioluminescence is the process by which living organisms convert chemical energy into photons of light and it is widely distributed throughout the animals, plants and fungi <ref>PMID:6358519</ref>. Species use bioluminescence as a survival tool in mating, defense and hunting. The one thing that all bioluminescent species have in common is that they all catalyze the reaction with an enzyme generically called a luciferase. All luciferases oxidize a substrate, which then decays back to the ground state while emitting a photon of light. This process is incredibly efficient with almost one photon of light produced per oxidation. While all lucifeases oxidize their substrates, the cofactors involved and reaction pathways used, vary widely <ref>PMID:775940</ref><ref>PMID:14444706</ref>. | Bioluminescence is the process by which living organisms convert chemical energy into photons of light and it is widely distributed throughout the animals, plants and fungi <ref>PMID:6358519</ref>. Species use bioluminescence as a survival tool in mating, defense and hunting. The one thing that all bioluminescent species have in common is that they all catalyze the reaction with an enzyme generically called a luciferase. All luciferases oxidize a substrate, which then decays back to the ground state while emitting a photon of light. This process is incredibly efficient with almost one photon of light produced per oxidation. While all lucifeases oxidize their substrates, the cofactors involved and reaction pathways used, vary widely <ref>PMID:775940</ref><ref>PMID:14444706</ref>. | ||
---- | ---- | ||
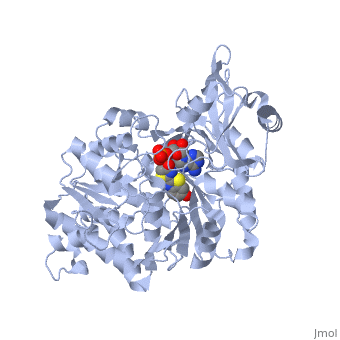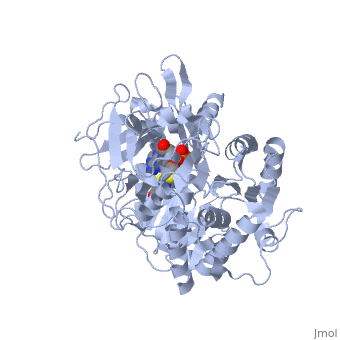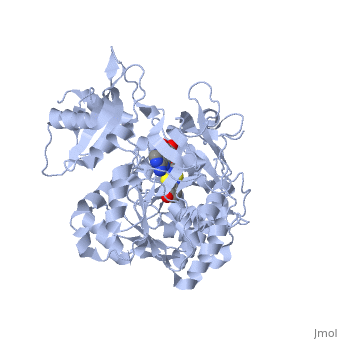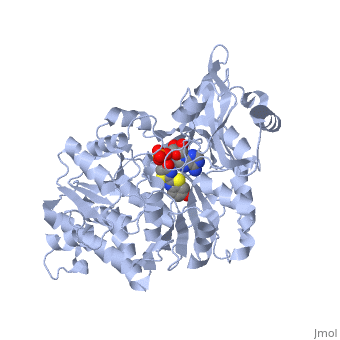
| + | <applet load='2d1r' size='400' frame='true' align='left' caption='Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP' /> | ||
The most studied of all luciferases is that of the Japanese <scene name='User:Matt_Whelihan/Rainbow_n-c/1'>Firefly</scene> ''Luciola cruciata''. This particular luciferase enzyme is located in the light emitting organ known as the lantern in the abdomen of the beetle. Firefly larvae glow green to ward off predators and adult fireflies use this mechanism of bioluminescence to attract mates. Luciferase binds ATP/Mg+ and D-luciferin and oxidizes it to Oxyluciferin with the products of one photon of yellow-green light, pyrophosphate, AMP and CO2<ref>PMID:775940</ref>. | The most studied of all luciferases is that of the Japanese <scene name='User:Matt_Whelihan/Rainbow_n-c/1'>Firefly</scene> ''Luciola cruciata''. This particular luciferase enzyme is located in the light emitting organ known as the lantern in the abdomen of the beetle. Firefly larvae glow green to ward off predators and adult fireflies use this mechanism of bioluminescence to attract mates. Luciferase binds ATP/Mg+ and D-luciferin and oxidizes it to Oxyluciferin with the products of one photon of yellow-green light, pyrophosphate, AMP and CO2<ref>PMID:775940</ref>. | ||
''L. cruciata'' luciferase is a 62 kDa monooxygenase with two distinct domains. The <scene name='User:Matt_Whelihan/Secondary_structure/1'>structure</scene> N-terminal domain (residues 4-436) is comprised of of an antiparalell β-barrel and two β-sheets, flanked by α-helices. The C-terminal domain (residues 440-544) is comprises of a separate α-β hinge. .<ref>PMID:8805533</ref> | ''L. cruciata'' luciferase is a 62 kDa monooxygenase with two distinct domains. The <scene name='User:Matt_Whelihan/Secondary_structure/1'>structure</scene> N-terminal domain (residues 4-436) is comprised of of an antiparalell β-barrel and two β-sheets, flanked by α-helices. The C-terminal domain (residues 440-544) is comprises of a separate α-β hinge. .<ref>PMID:8805533</ref> | ||
| Line 13: | Line 14: | ||
| - | <applet load='2d1r' size='400' frame='true' align='left' caption='Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP' /> | ||
---- | ---- | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Revision as of 03:19, 20 April 2009
2d1r
Japanese Firefly Luciferase
Bioluminescence is the process by which living organisms convert chemical energy into photons of light and it is widely distributed throughout the animals, plants and fungi [1]. Species use bioluminescence as a survival tool in mating, defense and hunting. The one thing that all bioluminescent species have in common is that they all catalyze the reaction with an enzyme generically called a luciferase. All luciferases oxidize a substrate, which then decays back to the ground state while emitting a photon of light. This process is incredibly efficient with almost one photon of light produced per oxidation. While all lucifeases oxidize their substrates, the cofactors involved and reaction pathways used, vary widely [2][3].
|
The most studied of all luciferases is that of the Japanese Luciola cruciata. This particular luciferase enzyme is located in the light emitting organ known as the lantern in the abdomen of the beetle. Firefly larvae glow green to ward off predators and adult fireflies use this mechanism of bioluminescence to attract mates. Luciferase binds ATP/Mg+ and D-luciferin and oxidizes it to Oxyluciferin with the products of one photon of yellow-green light, pyrophosphate, AMP and CO2[4]. L. cruciata luciferase is a 62 kDa monooxygenase with two distinct domains. The N-terminal domain (residues 4-436) is comprised of of an antiparalell β-barrel and two β-sheets, flanked by α-helices. The C-terminal domain (residues 440-544) is comprises of a separate α-β hinge. .[5] P. pyralis luciferase shares significant sequence and mechanistic homology with peptide synthetases and acylCoA ligases. These enzymes belong to a superfamily of adenylate-forming enzymes that catalyze activation reactions between ATP and a carboxyl group of their substrates. Despite high sequence homology, there are only seven residues that are conserved across this superfamily (Gly200, Lys206, Glu344, Asp422, Arg437, Gly446, and Glu455). These residues are believed to be integral to the binding of ATP and the formation of an adenylate compound. [6]. These seven highly conserved amino acids identify the which is located in the large cleft between the two adjacent N and C-terminal domains.
References
- ↑ Hastings JW. Biological diversity, chemical mechanisms, and the evolutionary origins of bioluminescent systems. J Mol Evol. 1983;19(5):309-21. PMID:6358519
- ↑ Deluca M. Firefly luciferase. Adv Enzymol Relat Areas Mol Biol. 1976;44:37-68. PMID:775940
- ↑ SELIGER HH, McELROY WD. Spectral emission and quantum yield of firefly bioluminescence. Arch Biochem Biophys. 1960 May;88:136-41. PMID:14444706
- ↑ Deluca M. Firefly luciferase. Adv Enzymol Relat Areas Mol Biol. 1976;44:37-68. PMID:775940
- ↑ Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ Turgay K, Krause M, Marahiel MA. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992 Sep;6(18):2743-4. PMID:1447981