User:Amy Rumora/Sandbox 1
From Proteopedia
| Line 15: | Line 15: | ||
An aspartic acid dyad is located in the active site between the N- and C- terminal lobes. Active site <scene name='User:Amy_Rumora/Sandbox_1/As_asp_greygreen/1'>Asp32 and Asp 228</scene> are in a coplanar configuration that allows them to coordinate and activate a water molecule. The activated water performs a nucleophilic attack on the peptide carbonyl group of the substrate. Hydrogen bonds between the carbonyl group of the aspartates and the substrate stabilize a geminal diol intermediate. A proton is then transferred from aspartate to the leaving group resulting in peptide bond breakage between Met671 and Asp672 of APP. The products of the hydrolysis reaction leave the active site. The active site of BACE1 is covered by a 10 amino acid flexible antiparallel β-hairpin called the <scene name='User:Amy_Rumora/Sandbox_1/Tyr71/1'>flap</scene>. This flap controls the entrance of substrates into the active site and is in closed conformation in the presence of bound inhibitors and open conformation in the inactive or apo structures. A Cα <scene name='User:Amy_Rumora/Sandbox_1/Apo_and_om99/1'>displacement</scene> of 7Å is visible in the inhibitor bound BACE1 complex in comparison to the apo structure (Patel). Tyrosine 71 is a conserved residue located at the tip of the flap that undergoes large conformational changes. In apo BACE1 structures, Tyr71 forms a hydrogen bond allowing the flap to adopt a more open conformation. In the presence of inhibitor, the phenolic ring of Tyr71 forms a weak hydrogen bond with Tryptophan 76 causing the flap to adopt a closed conformation. Additionally, the 10s loop undergoes conformational change in the presence of an inhibitor. | An aspartic acid dyad is located in the active site between the N- and C- terminal lobes. Active site <scene name='User:Amy_Rumora/Sandbox_1/As_asp_greygreen/1'>Asp32 and Asp 228</scene> are in a coplanar configuration that allows them to coordinate and activate a water molecule. The activated water performs a nucleophilic attack on the peptide carbonyl group of the substrate. Hydrogen bonds between the carbonyl group of the aspartates and the substrate stabilize a geminal diol intermediate. A proton is then transferred from aspartate to the leaving group resulting in peptide bond breakage between Met671 and Asp672 of APP. The products of the hydrolysis reaction leave the active site. The active site of BACE1 is covered by a 10 amino acid flexible antiparallel β-hairpin called the <scene name='User:Amy_Rumora/Sandbox_1/Tyr71/1'>flap</scene>. This flap controls the entrance of substrates into the active site and is in closed conformation in the presence of bound inhibitors and open conformation in the inactive or apo structures. A Cα <scene name='User:Amy_Rumora/Sandbox_1/Apo_and_om99/1'>displacement</scene> of 7Å is visible in the inhibitor bound BACE1 complex in comparison to the apo structure (Patel). Tyrosine 71 is a conserved residue located at the tip of the flap that undergoes large conformational changes. In apo BACE1 structures, Tyr71 forms a hydrogen bond allowing the flap to adopt a more open conformation. In the presence of inhibitor, the phenolic ring of Tyr71 forms a weak hydrogen bond with Tryptophan 76 causing the flap to adopt a closed conformation. Additionally, the 10s loop undergoes conformational change in the presence of an inhibitor. | ||
| - | [[Image:Asparticmech. | + | [[Image:Asparticmech.png]] |
http://en.wikipedia.org | http://en.wikipedia.org | ||
Revision as of 07:07, 23 April 2009
Contents |
BACE1
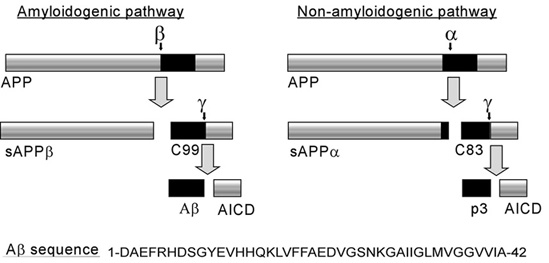
BACE1 (β-site of APP cleaving enzyme) also called β-Secretase and memapsin-2 is a 52 kD class I transmembrane aspartic acid protease that cleaves the Amyloid Precursor Protein (APP) in a rate limiting step that contributes to the accumulation of β-amyloid plaques in Alzheimer’s disease (AD). A subsequent cleavage by γ-secretase generates a 40 or 42 amino acid β-amyloid peptide. These peptides can form Aβ plaques that may have deleterious effects on neuronal function and contribute to pathologies of AD. Under normal conditions, BACE1 activity generates a monomeric and soluble Aβ peptide that may play a physiological role in decreasing excitotoxicity [1] and neurotransmission at glutamatergic synapses. Additionally, α-secretase and γ-secretase cleave APP to generate p3 and the carboxy terminal fragment AICD in a non-amyloidogenic pathway. In AD, amyloidogenic pathways become preferential over non-amyloidogenic and Aβ plaques appear under increased levels of BACE1 catalytic activity.
Overall structure of BACE1
| |||||||||
| 1fkn, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | |||||||||
| Activity: | Memapsin 2, with EC number 3.4.23.46 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
BACE1 is a bilobal protein that consists of luminal N-lobe and C-lobe anchored to the plasma membrane by a transmembrane domain that exposes a small cytosolic domain. The catalytic core of BACE1, shown here, spans amino acid residues 43-385. BACE1 is a member of the mainly beta class of proteins with a β-barrel architecture. This structure is composed of (shown in green and grey, respectively) (science 2000). Unlike other class I aspartic proteases, insertions and an extension in the C-terminus of BACE1 creates an enlarged C-terminal lobe. are localized on the surface of the C-lobe adjacent from the N-terminus of bound inhibitors. Four acidic amino acid residues on the F insertion loop form a negatively charged area on the surface of the enzyme. are located on the surface of the C-lobe near the C-terminus of bound inhibitors. These BACE1 insertions may function to form interactions with cell surface components and serve to enlarge the C-terminal surface area. A 35 residue C-terminal (359-385) forms an ordered β structure spanning residues 369-376 and a helix between residues 378-383. This structure may form a stem with the transmembrane domain. BACE1 contains three disulfide bonds that vary from other Class I aspartic proteases there are three structural disulfide bonds These structural disulfide bonds are important for maintaining the correct fold of BACE1
The C-terminus participates in protein-protein interactions and undergoes post-translational modification while the N-terminus contains catalytic aspartic acid residues that become active under acidic conditions of the endosome or golgi. Additionally, the distribution of disulfide bridges in BACE1 varies from the evolutionarily conserved. BACE1 contains three disulfide bonds there are three structural disulfide bonds These structural disulfide bonds are important for maintaining the correct fold of BACE1
Structural elements involved in catalysis
| |||||||||
| 1w50, resolution 1.75Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Activity: | Memapsin 2, with EC number 3.4.23.46 | ||||||||
| Related: | 1fkn, 1m4h, 1py1, 1sgz, 1ujj, 1ujk, 1w51 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
An aspartic acid dyad is located in the active site between the N- and C- terminal lobes. Active site are in a coplanar configuration that allows them to coordinate and activate a water molecule. The activated water performs a nucleophilic attack on the peptide carbonyl group of the substrate. Hydrogen bonds between the carbonyl group of the aspartates and the substrate stabilize a geminal diol intermediate. A proton is then transferred from aspartate to the leaving group resulting in peptide bond breakage between Met671 and Asp672 of APP. The products of the hydrolysis reaction leave the active site. The active site of BACE1 is covered by a 10 amino acid flexible antiparallel β-hairpin called the . This flap controls the entrance of substrates into the active site and is in closed conformation in the presence of bound inhibitors and open conformation in the inactive or apo structures. A Cα of 7Å is visible in the inhibitor bound BACE1 complex in comparison to the apo structure (Patel). Tyrosine 71 is a conserved residue located at the tip of the flap that undergoes large conformational changes. In apo BACE1 structures, Tyr71 forms a hydrogen bond allowing the flap to adopt a more open conformation. In the presence of inhibitor, the phenolic ring of Tyr71 forms a weak hydrogen bond with Tryptophan 76 causing the flap to adopt a closed conformation. Additionally, the 10s loop undergoes conformational change in the presence of an inhibitor.
Image:Asparticmech.png http://en.wikipedia.org
BACE1- a therapeutic target for AD
| |||||||||
| 2zhr, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | |||||||||
| Gene: | BACE1 (Homo sapiens) | ||||||||
| Activity: | Memapsin 2, with EC number 3.4.23.46 | ||||||||
| Related: | 2zhs, 2zht, 2zhu, 2zhv | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
BACE1 is a major therapeutic target due it's role in generating Aβ plaques that contribute to AD pathogenesis. There are currently over 70 structures of BACE1 complexed with inhibitors in the protein data bank http://www.rcsb.org/. These studies have not only developed inhibitors that efficiently block BACE1 activity, but also provide a greater understanding of the residues and regions that interact with the substrate in the active site of BACE1. The first inhibitor crystallized in complex with BACE1 was OM99-2. is a transition state analogue inhibitor that is eight residues in length (Glu-Val-Asn-Leu*-Ala-Ala-Glu-Phe) designated P4-P4’ (P4-P3-P2-P1*-P1’-P2’-P3’-P4’). OM99-2 inhibits BACE1 cleavage between the P1*-P1’ bond because of the presence of a hydroxyethylene isotere (*) that cannot be cleaved. The S1 and S3 sites that bind P1 and P3 are hydrophobic pockets that bind uncharged residues of the substrate (understand). On the other hand, sites S2 and S4 contain hydrophilic residues such as Arg that accommodate charged residues (39, 49 understand). Sites S5-S7 are in the proximity of the insertion helix and may be important for recognition of the substrate (S5, S6, S7 cite). The maintains a closed conformation over OM99-2 through hydrogen bonding between the conserved Tyr71 residue and the substrate at positions P1 and P2' (understand 47). Additionally, a conformational displacement is observed for the in the S3 region of the BACE1 active site.
Evolutionary conservation among asparic proteases
BACE1 is evolutionarily related to other enzymes in the aspartic protease family. Pepsin, Cathepsin D, Chymosin, and renin fall into the aspartic protease family and are all evolutionarily related to BACE1. Aside from the conserved aspartate residues that catalyze the hydrolysis of the peptide bond in APP, there are several other key structural features that are conserved across this family.
Links
- 1w50 (Apo structure of bace (beta secretase))
- 1w51 (Bace (beta secretase) in complex with a nanomolar non-peptidic inhibitor)
- 2zhs, 2zht, 2zhu, 2zhv (Crystal structure of BACE1 at varying pH)
- 2zhr (Crystal structure of BACE1 in complex with OM99-2 at pH 5.0)
- 1fkn (Structure of Beta-Secretase Complexed with Inhibitor)
References