Factor IX
From Proteopedia
| Line 100: | Line 100: | ||
-------------- | -------------- | ||
http://www.natuurlijkerwijs.com/english/ ]] | http://www.natuurlijkerwijs.com/english/ ]] | ||
| - | |||
| - | |||
| Line 119: | Line 117: | ||
'''Step 3 and 4:''' Active vitamin K in the hydroquinone form is regenerated in two reaction step | '''Step 3 and 4:''' Active vitamin K in the hydroquinone form is regenerated in two reaction step | ||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 21:10, 26 April 2009
-----This page is still under construction-------
|
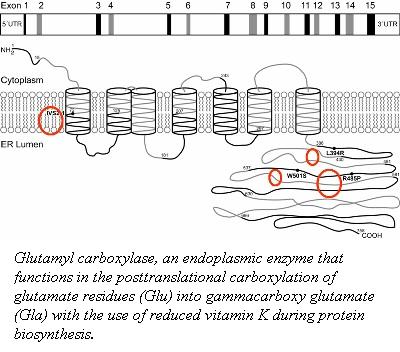
Factor IX (plasma thromboplastin component, Christmas factor, or hemophilia B factor) is a 57-kDa vitamin K-dependent procoagulant glycoprotein. It is synthesized by the liver hepatocyte as a pre-prozymogen that requires extensive posttranslational modification. The pre-prozymogen contains a pre-peptide (hydrophobic signal peptide) at its amino terminal that transports the growing polypeptide into the lumen of the Endoplasmic Reticulum. Once inside the ER, this signal peptide is cleaved by a signal peptidase. A pro-peptide functions as a recognition element for a vitamin K-dependent carboxylase (γ-glutamyl carboxylase) which modifies 12 glutamic acid residues to gamma-carboxyglutamyl ( ) residues. These residues are required for the association with the anionic phospholipid surface through Ca2+-dependent binding. The Domain is followed by two epidermal growth factor domains ( and ). The N-terminus of contains a Ca2+ binding site, while the C-terminus connects to a hydrophobic pocket of and a salt bridge through Lys122 ( residue) and Gln74 (). connects to the domain through a linker peptide and is required for a proper orientation and folding of . To have a physiologically active factor IX, two cleaves must occur to remove a 35 amino acid region that precedes the catalytic region. The first cleave is at Arg145, generating an inactive FIXα. The second cleavage is at Arg180 results in a catalytically active molecule FIXaβ. This resulting heterodimer is held by a disulfide bridge at Cys132-Cys289. The contains a catalytic triad of . Upon cleave at Arg180, Val181 can form a salt bridge with Asp364, which is a characteristic of active . The active FIXa, can then interact with its cofactor, FVIIIa, to form a membrane-bound Xase complex, which activated FX to FXa.
Contents |
Gene Structure and Expression:
The gene for factor IX is located on the long arm of chromosome X between positions 26.3- and 27.1 and contains eight exons and seven introns, which segregate the FIX gene into specific structural regions.
| I | II | III | IV | V | VI | VII | VIII |
Exon I encodes the hydrophobic signal peptide that targets the FIX into the lumen of the Endoplasmic Reticulum. Exon II codes for the pro-peptide and the Gla domain. Exon III encodes the , that inserts itself into the lipid membrane, anchoring FIX. Exon IV and V, code for EGF-1 and EGF-2, the serine protease region is encoded by exon VI-VIII.
γ-Carboxyglutamate: Post-translational carboxylation of glutamate residues (Glu) into gammacarboxy glutamate (Gla)
The post-translational modification of the glutamic acid residues of the FIX polypeptide is carried out by vitamin K-dependent γ-glutamyl carboxylase (1). Vitamin K-dependent carboxylase is a membrane associated protein in the endoplasmic reticulum. It converts a multiple of glutamic acid residues which are located within 40 residues of a propeptide-containing sequence into γ-carboxyglutamate (2).
The association ofvitamin K-dependent carboxylase and its substrates is dependent on the 18-amino acid propeptide sequence (3, 4). The propeptide anchors the substrate to the carboxylase for a carboxylation reaction to occur. This binding of the propeptide to carboxylase stimulates the incorporation of carbon dioxide into the glutamate residues with the use of a reduced vitamin K and oxygen (4, 5). The residues 495-513 of the carboxylase function as an internal propeptide that is homologous to the the propeptide sequence.
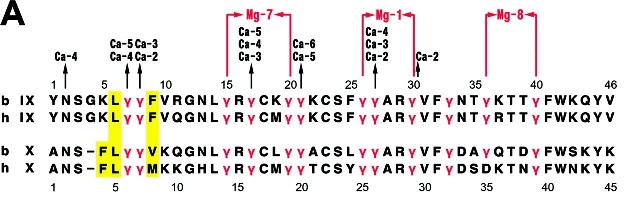
To identify the internal propetide region of the Vitamin K-dependent carboxylase, five mutant carboxylase molecules (F496A, V502A, Q503R, Q503N, and P504Q) were generated identify the residues responsible for homologous substrate propeptide binding.
The alignment of the proposed internal propeptide region with the consensus human propeptide and the corresponding region of the mutations used. Red residues represent mutations that make the internal propeptide less like the consensus sequence. Residues mutated to be more like the consensus sequence are shown in Blue.
| Consensus | AVFLS2EQANOVLQRRRR |
| Internal_Propep | PFQRTSWVQPLLNDLSPW |
| F496A | PAQRTSWVQPLLNDLSPW |
| P495V / Q497L | VFLRTSWVQPLLNDLSPW |
| V502A | PFQRTSWAQPLLNDLSPW |
| V502A /P504Q | PFQRTSWAQQLLNDLSPW |
| Q503N | PFQRTSWVNPLLNDLSPW |
| Q503R | PFQRTSWVRPLLNDLSPW |
| P504Q | PFQRTSWVQQLLNDLSPW |
Mutations:
Phe496 correspond to residue 16: Consensus phenylalanine to alanine leads to a reduced affinity of the internal propeptide for the propeptide binding site.
Val502 corresponds to position 10, Consensus val to alanine, leads to an increase in the affinity of the proposed internal propeptide for the propeptide binding site.
Glutamine 503 corresponds to position 9, Consensus Gln to Arg leads to a reduced affinity of the internal propeptide for the propeptide binding site. Consensus Gln to Asn, leads the consensus residue to have a modest increase in the apparent affinity.
Pro 504 corresponds to position 8, not a highly conserved position; however, the propeptide may have an -helical structure (6), and in this case the proline will disrupt the substrate propeptide helix. Changing proline 504 to the consensus sequence residue, glutamine, has a small increase in the affinity of the internal propeptide for its binding site.
These mutations identified that residues 495-513 in the carboxylase act as an internal propeptide binding site (7).
γ-Carboxylation Reaction
http://www.natuurlijkerwijs.com/english/
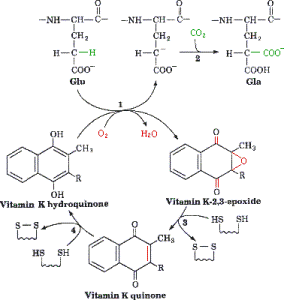
γ-Carboxylation Reaction is catalyzed by the γ-glutamyl carboxylase in the ER and requires a reduced form of vitamin K, oxygen, and carbon dioxide. The reaction is initiated by the removal of a hydrogen atom at the γ position of glutamate. This reaction creates a carbon ion that reacts with carbon dioxide thus forming γ-carboxy glutamic acid. As the protein is carboxylated using carbon dioxide the reduced vitamin K (hydroquinone) is oxidized to epoxide.
In order for this reaction to continue vitamin K must be regenerated, which is carried out by nicotinamide phosphate, thus allowing the continuation of the cycle. Protein carboxylation, provides a region which allows the protein to associate with the membrane anionic phospholipid surface, thus allowing close proximity to other components of coagulation. Without vitamin K, coagulation precursor proteins would circulate through the plasma, but would have minimal function.
Step 1: Vitamin K is in its active hydroquinone form: In this reaction a proton (H+ ion) is removed from the glutamic acid in an oxygen (O2) consuming reaction. This reaction leads to a carbon ion aring on the glutamic acid molecule.
Step 2: The glutamic acid carbon ion reacts with CO2 forming gamma-carboxyglutamate.
Step 3 and 4: Active vitamin K in the hydroquinone form is regenerated in two reaction step
γ-Carboxyglutamic Acid (Gla) Domain
The Gla domain is situated at the N-terminus of coagulation factor IX, and is evolutionary conserved in other vitamin K dependent proteins such as factor VII, X, and prothrombin. A Gla domain is made up of 10-13 of γ-carboxyglutamic acid residues and requires both Ca2+ and Mg2+ ions for membrane association and stabilization of its active three dimensional conformation (2-4).
|
|
Proteopedia Page Contributors and Editors (what is this?)
Nadia Dorochko, Michal Harel, Alexander Berchansky, David Canner