User:Maitreyee Mukherjee/Sandbox 1
From Proteopedia
| Line 3: | Line 3: | ||
'''<font color='red'>INTRODUCTION:</font>''' | '''<font color='red'>INTRODUCTION:</font>''' | ||
| - | ''Rhodobacter sphaeroides'' is a metabolically versatile purple photosynthetic α-proteobacteria<ref>PMID: 14999403</ref> which produces polyhydroxybutyrate (PHB) as inclusions inside its cell. PHB is a biopolymer which has the properties similar to synthetic polymers such as polyethylene and hence has a potential of being used as a biopolymer. The biochemical pathway for PHB production in this organism is complex and is controlled by the actions of several gene products<ref>PMID:10416645</ref>. PHB synthase encoded by ''phaC'' is responsible for conversion of the monomeric precursor of PHB R(-)-β-Hydroxybutyryl-CoA into polyhydroxybutyrate | + | ''Rhodobacter sphaeroides'' is a metabolically versatile purple photosynthetic α-proteobacteria<ref>PMID: 14999403</ref> which produces polyhydroxybutyrate (PHB) as inclusions inside its cell. PHB is a biopolymer which has the properties similar to synthetic polymers such as polyethylene and hence has a potential of being used as a biopolymer. The biochemical pathway for PHB production in this organism is complex and is controlled by the actions of several gene products<ref>PMID:10416645</ref>. PHB synthase encoded by ''phaC'' is responsible for conversion of the monomeric precursor of PHB R(-)-β-Hydroxybutyryl-CoA into polyhydroxybutyrate <ref> Hustede, E., and A. Steinbuchel. 1993. Characterization of the polyhydroxyalkanoate synthase gene locus of Rhodobacter sphaeroides. Biotechnology Letters. 15:709–714.</ref>.Rhodobacter sphaeroides possesses the Class I phylogenetic group of PHA synthases<ref>PMID:14620841</ref>. Class I PHA synthases prefer utilization of CoA thioesters of various (R)-3-hydroxy fatty acids comprising of 3 to 5 carbon atoms<ref>PMID:12954080</ref>. |
'''<font color='red'>FUNCTION:</font>''' | '''<font color='red'>FUNCTION:</font>''' | ||
| Line 135: | Line 135: | ||
1. In ''R.sphaeroides'' FJ1, ''phbC'' has been found to be present on one of two gene locus identified to be involved in the production of PHB. The gene ''phbC'' is present on the same locus along with the other genes responsible for PHB syn thesis such as ''phbZ'', ''phbP'' and ''phbR''. <ref>PMID: 16440119</ref> | 1. In ''R.sphaeroides'' FJ1, ''phbC'' has been found to be present on one of two gene locus identified to be involved in the production of PHB. The gene ''phbC'' is present on the same locus along with the other genes responsible for PHB syn thesis such as ''phbZ'', ''phbP'' and ''phbR''. <ref>PMID: 16440119</ref> | ||
| + | |||
| + | 2. In a paper by Fales et.al. in 2001, the investigators talk about a possible connection between PHB synthase activity and ''hemA'' transcription in ''R.sphaeroides''. The investigators also present a schematic diagram of a proposed dual pathway of PHB synthesis in this organism.<ref>PMID: 11160087</ref> No further investigations have been published regarding this untill now. | ||
| + | |||
<font color='red'>'''PROBABLE RESEARCH:FOR BETTER UNDERSTANDING OF THIS PROTEIN:CHALLENGE YOUR UNDERSTANDING'''</font> | <font color='red'>'''PROBABLE RESEARCH:FOR BETTER UNDERSTANDING OF THIS PROTEIN:CHALLENGE YOUR UNDERSTANDING'''</font> | ||
Revision as of 23:16, 26 April 2009
|
INTRODUCTION:
Rhodobacter sphaeroides is a metabolically versatile purple photosynthetic α-proteobacteria[1] which produces polyhydroxybutyrate (PHB) as inclusions inside its cell. PHB is a biopolymer which has the properties similar to synthetic polymers such as polyethylene and hence has a potential of being used as a biopolymer. The biochemical pathway for PHB production in this organism is complex and is controlled by the actions of several gene products[2]. PHB synthase encoded by phaC is responsible for conversion of the monomeric precursor of PHB R(-)-β-Hydroxybutyryl-CoA into polyhydroxybutyrate [3].Rhodobacter sphaeroides possesses the Class I phylogenetic group of PHA synthases[4]. Class I PHA synthases prefer utilization of CoA thioesters of various (R)-3-hydroxy fatty acids comprising of 3 to 5 carbon atoms[5].
FUNCTION:
The enzyme catalyses the conversion of the monomeric precursor of PHB into PHB with the release of CoA. In other words these enzymes mediate the conversion of a soluble substrate into polymerized insoluble inclusions inside the cells of these bacteria. Upon covalent catalysis of polyester chain formation, this soluble enzyme gets converted to amphipathic enzyme (The amphipathic enzyme conjugates are analogous to certain membrane-bound proteins such as cytochrome b, which have been shown to comprise a hydrophilic protein core anchored to a lipid bilayer by a hydrophobic polypeptide tail). Initiation of a self assembly process occurs and this results in the formation of the insoluble cytoplasmic inclusions with a phospholipids monolayer and covalently attached polyester synthases at the surface.
PROPERTIES:
| N | C |
| Amino Terminus | Carboxy Terminus |
| 5' | 3' |
| N, 5' | C, 3' |
Hydrophobic, Polar
Alpha Helices, Beta Strands , Turns.
Number of amino acids: 601
Molecular weight: 66831.6
Theoretical pI: 5.63
Amino acid sequence:
1 mateeqspgs grdaqferln anltridels krltaaltkr klsdpalhgp sgdvflkamt
61 aymaemmqnp akilehqisf wgkslkhyve aqhqlvkgel kpppdvtpkd rrfsnplwqt
121 hpffnylkqq ylmnaeavnq avealehiep sdkkrveyfs rqivdlfspt nffgtnpdal
181 eraiatdges lvqglenlvr dieanngdll vtladpeafq vgqnlatteg svvyrnrmfe
241 liqykpttet vhetpllifp pwinkfyild lkpqnsllkw lvdqgftvfv vswvnpdksy
301 agigmddyir egymramaev rsitrqkqin avgyciagtt ltltlahlqk agdpsvrsat
361 ffttltdfsd pgevgvflnd dfvdgierqv avdgildktf msrtfsylrs ndliyqpaik
421 symmgeappa fdllywngdg tnlpaqmave ylrglcqqdr laggtfpvlg spvglkdvtl
481 pvcaiacetd hiapwkssfn gfrqfgstdk tfilsqsghv agivnppsrn kyghytnegp
541 agtpesfreg aefhagswwp rwgawlaers gkqvparqpg dskhpelapa pgsyvaavgg
601 a
Amino acid Composition:
Ala (A) 52 8.7%, Arg (R) 29 4.8%, Asn (N) 26 4.3%, Asp (D) 34 5.7%, Cys (C) 4 0.7%, Gln (Q) 29 4.8%, Glu (E) 35 5.8%, Gly (G) 46 7.7%, His (H) 13 2.2%, Ile (I) 24 4.0%, Leu (L) 54 9.0%, Lys (K) 28 4.7%, Met (M) 14 2.3%, Phe (F) 31 5.2%, Pro (P) 40 6.7%, Ser (S) 36 6.0%, Thr (T) 37 6.2%, Trp (W) 11 1.8%, Tyr (Y) 20 3.3%. Val (V) 38 6.3% Pyl (O) 0 0.0% Sec (U) 0 0.0%
Total number of negatively charged residues (Asp + Glu): 69 Total number of positively charged residues (Arg + Lys): 57
Atomic composition:
Carbon C 3004 Hydrogen H 4612 Nitrogen N 808 Oxygen O 888 Sulfur S 18
Total number of atoms: 9330
Extinction coefficients:
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient 90550 Abs 0.1% (=1 g/l) 1.355, assuming ALL Cys residues appear as half cystines Ext. coefficient 90300 Abs 0.1% (=1 g/l) 1.351, assuming NO Cys residues appear as half cystines
Estimated half-life:
The N-terminal of the sequence considered is M (Met).
The estimated half-life is: 30 hours (mammalian reticulocytes, in vitro). >20 hours (yeast, in vivo). >10 hours (Escherichia coli, in vivo).
Instability index:
The instability index (II) is 41.97 This classifies the protein as unstable.
Aliphatic index: 77.60
Grand average of hydropathicity (GRAVY): -0.330
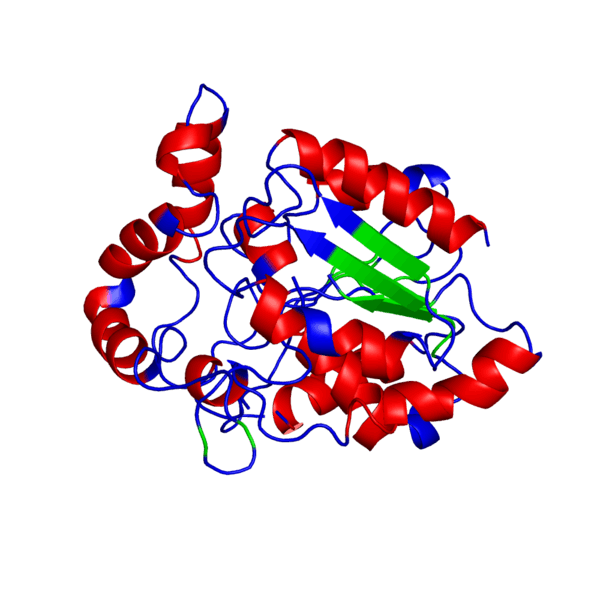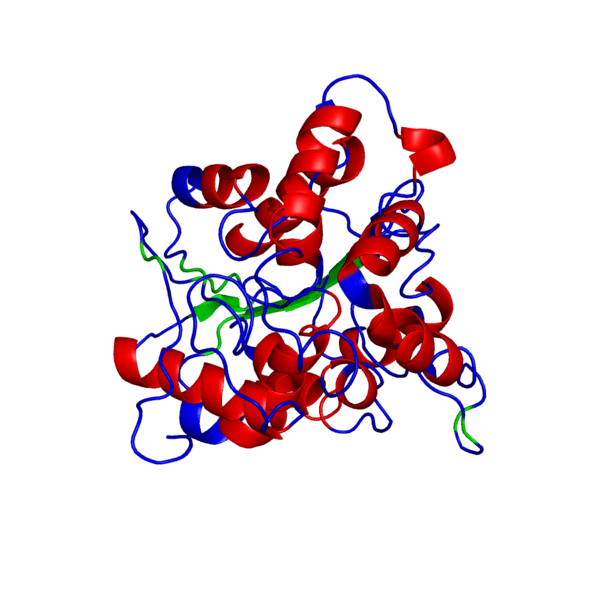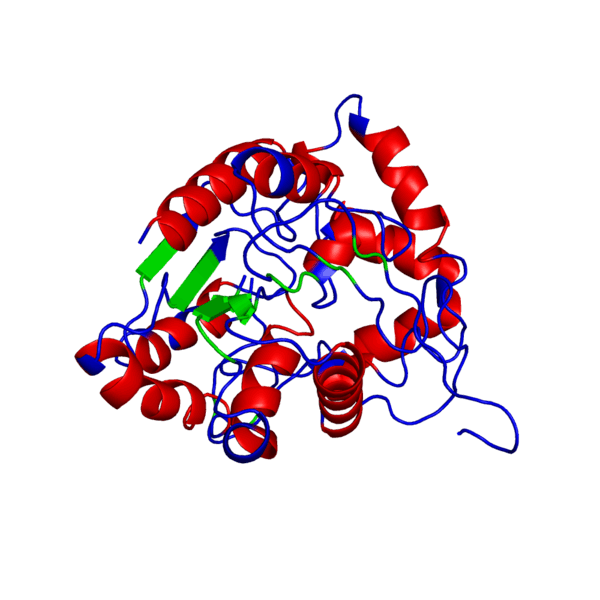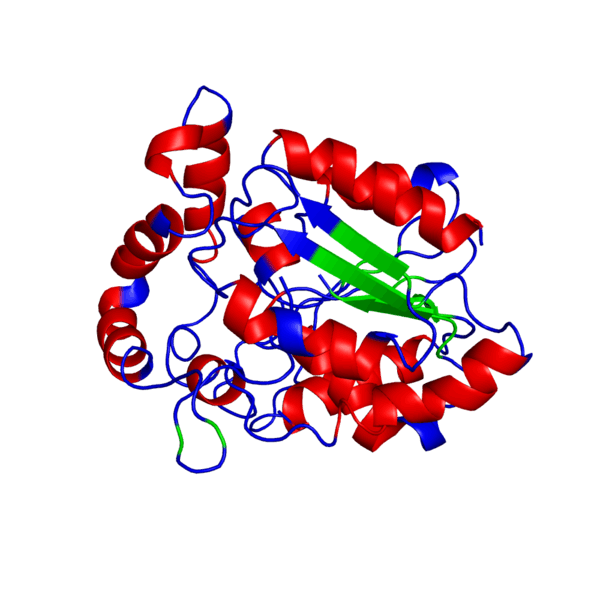
ANIMATED IMAGE OF THE PUTATIVE PHB SYNTHASE DOMAIN SHOWING AMINO ACIDS 286 TO 547 IN Rhodobacter sphaeroides OBTAINED FROM POLYVIEW-3D
CURRENT RESEARCH: KNOWLEDGE OF THIS PROTEIN IN R.sphaeroides AS WELL AS OTHER ORGANISMS:
1. In R.sphaeroides FJ1, phbC has been found to be present on one of two gene locus identified to be involved in the production of PHB. The gene phbC is present on the same locus along with the other genes responsible for PHB syn thesis such as phbZ, phbP and phbR. [6]
2. In a paper by Fales et.al. in 2001, the investigators talk about a possible connection between PHB synthase activity and hemA transcription in R.sphaeroides. The investigators also present a schematic diagram of a proposed dual pathway of PHB synthesis in this organism.[7] No further investigations have been published regarding this untill now.
PROBABLE RESEARCH:FOR BETTER UNDERSTANDING OF THIS PROTEIN:CHALLENGE YOUR UNDERSTANDING
1. What is the molecular basis of regulation of PHB synthesis in this organism and what is the role of this protein in it?
2. How does environmental factors such as presence or absence of oxygen or nitrogen relate to the activity of this protein in R.sphaeroides?
3. What are the activators or repressor factors, if any, responsible for production of this protein in this organism?
HOW WAS THE ANIMATED IMAGE GENERATED?
1. Go to the POLYVIEW 3D homepage, http://polyview.cchmc.org/polyview3d.html
2. On the submission form, first select 'animation' in the "type of request" section, select the size of the animation to be generated in pixels(here the size is 600), then upload the PDB format protein structure file in the "source of structural data" section.
3. On the "chain color and rendering section" select 'cartoon' and 'secondary structure'.
4. On "advanced structural annotation" section select 'docking models in Capri format'.
5. Any other forms for the animation may be selected by referring to the "Samples" according to the protein structure to be animated.
HOW WAS THE JMOL IMAGE GENERATED?
1. First retrieve your protein sequence from http://www.ncbi.nlm.nih.gov/.
2. Go to 3D-JIGSAW page http://bmm.cancerresearchuk.org/~3djigsaw/ and paste the sequence on the submission page. A .pdb format image of your protein will be sent to you on your email which can be opened by RASMOL.
3. Upload this file on Proteopedia and then load the JMol applet for the protein following instructions on the Help:Editing page http://www.proteopedia.org/wiki/index.php/Help:Editing.
4. You can edit your protein by using the scene authoring tools after loading the applet.
REFERENCES:
- ↑ Zeilstra-Ryalls JH, Kaplan S. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell Mol Life Sci. 2004 Feb;61(4):417-36. PMID:14999403 doi:10.1007/s00018-003-3242-1
- ↑ Rehm BH, Steinbuchel A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol. 1999 Jun-Jul;25(1-3):3-19. PMID:10416645
- ↑ Hustede, E., and A. Steinbuchel. 1993. Characterization of the polyhydroxyalkanoate synthase gene locus of Rhodobacter sphaeroides. Biotechnology Letters. 15:709–714.
- ↑ Stubbe J, Tian J. Polyhydroxyalkanoate (PHA) hemeostasis: the role of PHA synthase. Nat Prod Rep. 2003 Oct;20(5):445-57. PMID:14620841
- ↑ Rehm BH. Polyester synthases: natural catalysts for plastics. Biochem J. 2003 Nov 15;376(Pt 1):15-33. PMID:12954080 doi:http://dx.doi.org/10.1042/BJ20031254
- ↑ Yang MK, Lin YC, Shen CH. Identification of two gene loci involved in poly-beta-hydroxybutyrate production in Rhodobacter sphaeroides FJ1. J Microbiol Immunol Infect. 2006 Feb;39(1):18-27. PMID:16440119
- ↑ Fales L, Kryszak L, Zeilstra-Ryalls J. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: effect of a transposon insertion in the hbdA gene. J Bacteriol. 2001 Mar;183(5):1568-76. PMID:11160087 doi:10.1128/JB.183.5.1568-1576.2001
1. Fales, L., L. Kryszak., and J. Zeilstra-Ryalls. 2001. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: effect of a transposon insertion in the hbdA gene. Journal of Bacteriology. 183:1568–1576.
2. Hustede, E., and A. Steinbuchel. 1993. Characterization of the polyhydroxyalkanoate synthase gene locus of Rhodobacter sphaeroides. Biotechnology Letters. 15:709–714.
3. Kolibachuk, D., A. Miller., and D. Dennis. 1999. Cloning, molecular analysis, and expression of the polyhydroxyalkanoic acid synthase (phaC) gene from Chromobacterium violaceum. Applied and Environmental Microbiology. 65:3561–3565.
4. Kranz, R.G., K. K. Gabbert., T. A. Locke., and M. T. Madigan. 1997. Polyhydroxyalkanoate production in Rhodobacter capsulatus: genes, mutants, expression, and physiology. Applied and Environmental Microbiology. 63:3003–3009.
5. Peoples, P.O., and A. J. Sinskey. 1989. Poly-ß-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16, identification and characterization of the PHB polymerase gene (phbC). The Journal of Biological Chemistry. 264(26):15298-15303.
6. Rehm, H. A. B., and A. Steinbüchel. 1999. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. International Journal of Biological Macromolecules. 25:3-19.
7. Rehm, Bernd H. A. B. 2003. Polyester synthases: natural catalysts for plastics. Biochemical Journal. 376:15–33.
8. Stubbe, J. A., and J. Tian. 2003. Polyhydroxyalkanoate (PHA) homeostasis: the role of PHA synthase. Natural Product Reports. 20:445-457.
9. Tae-Kwon, K. J. Young-Mi. M. Tri Vo., S.Suteaki., and L.Yong-Hyun. 2006. Metabolic engineering and characterization of phaC1 and phaC2 genes from Pseudomonas putida KCTC1639 for overproduction of medium-chain-length polyhydroxyalkanoate. Biotechnology Progress.22: 1541-1546.
10. Ueda, S., T. Yabutani., A. Maehara., and T. Yamane. 1996. Molecular analysis of the poly(3-hydroxyalkanoate) synthase gene from a methylotrophic bacterium, Paracoccus denitrificans. Journal of Bacteriology. 178:774–779.