User:Yana Fedotova/Sandbox 1
From Proteopedia
| Line 7: | Line 7: | ||
== Structure of the Proposed PrrA Protein == | == Structure of the Proposed PrrA Protein == | ||
''<applet load='PrrA.pdb' size='300' color='color' frame='true' align='right' caption='3D Image of proposed PrrA protein'/> | ''<applet load='PrrA.pdb' size='300' color='color' frame='true' align='right' caption='3D Image of proposed PrrA protein'/> | ||
| - | <scene name='1umq/Prra_model/4'>PrrA Model</scene> | ||
from http://ca.expasy.org/tools/protparam.html | from http://ca.expasy.org/tools/protparam.html | ||
| Line 310: | Line 309: | ||
*Page Edited by Yana Fedotova, email: yana.fedotova@gmail.com, yfedot@bgsu.edu | *Page Edited by Yana Fedotova, email: yana.fedotova@gmail.com, yfedot@bgsu.edu | ||
*Acknowledgment to Dr. Jill Zeilstra-Ryalls, Susana Retamal and Adam Meade | *Acknowledgment to Dr. Jill Zeilstra-Ryalls, Susana Retamal and Adam Meade | ||
| + | |||
| + | <scene name='1umq/Prra_model/4'>PrrA Model</scene> | ||
Revision as of 02:45, 1 May 2009
Contents |
Photosynthesis Response Regulator, PrrA
Background Information
PrrA is a DNA binding protein and a essential part of the two-component signal transduction regulatory system, PrrBA. Rhodobacter capsulatus contains RegA considered a homologue to PrrA protein in R. sphaeroides. PrrA is proposed to be a master regulator/response regulator involved in oxygen regulation of photosynthesis genes expression responding to changes to the rate of electron flow as means to maintain redox balance, such as a balance between energy production and energy consumption in the cell (4). PrrA is thought to play an important function in carbon dioxide and nitrogen fixations, photosynthesis as well as proton oxidation and uptake (1). PrrB is considered a sensor histidine kinase bound to the cytoplasmic membrane that can phosphorylate PrrA when it senses changes the aforementioned changes in the cell. Interestingly, “1,058 genes were proposed to be regulated out of 4,284 genes as represented on the GeneChip of R. sphaeroides 2.4.1” by PrrA (2).
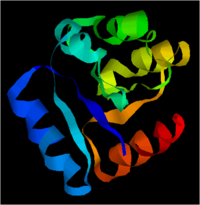
Structure of the Proposed PrrA Protein
|
from http://ca.expasy.org/tools/protparam.html
Molecular weight: 20483.5 Da
Number of Amino Acids: 184
Theoretical pI: 6.85
Estimated half-life:
The N-terminal of the sequence considered is M (Met).
The estimated half-life is:
- 30 hours (mammalian reticulocytes, in vitro).
- >20 hours (yeast, in vivo).
- >10 hours (Escherichia coli, in vivo).
Total number of negatively charged residues (Asp + Glu): 28
Total number of positively charged residues (Arg + Lys): 28
Atomic composition: Carbon C 894 Hydrogen H 1468 Nitrogen N 268 Oxygen O 268 Sulfur S 7
Total number of atoms: 2905
Extinction coefficients:
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient 11585
Abs 0.1% (=1 g/l) 0.566, assuming ALL Cys residues appear as half cystines
Ext. coefficient 11460
Abs 0.1% (=1 g/l) 0.559, assuming NO Cys residues appear as half cystines
Instability index: 55.68
Aliphatic index: 95.49
Grand Average of Hydropathicity (GRAVY): -0.311
Alpha-helices: 34.78%
Extended (Beta-sheets): 20.65%
Other (Loops): 44.57%
Amino Acid Sequence of PrrA Fragment
1 maedlvfelg adrslllvdd depflkrlak amekrgfvle taqsvaegka iaqarppaya 61 vvdlrledgn gldvvevlre rrpdcrivvl tgygaiatav aavkigatdy lskpadanev 121 thallakges lppppenpms adrvrwehiq riyemcdrnv setarrlnmh rrtlqrilak 181 rspr
| Amino Acid | Number present | Percentage of total present |
|---|---|---|
| Ala (A) | 15 | 13.0% |
| Arg (R) | 10 | 10.9% |
| Asn (N) | 6 | 2.7% |
| Asp (D) | 10 | 7.1% |
| Cys (C) | 1 | 1.1% |
| Gln (Q) | 5 | 2.2% |
| Glu (E) | 11 | 8.2% |
| Gly (G) | 9 | 4.9% |
| His (H) | 10 | 1.6% |
| Ile (I) | 3 | 3.8% |
| Leu (L) | 21 | 10.9% |
| Lys (K) | 6 | 4.3% |
| Met (M) | 6 | 2.7% |
| Phe (F) | 2 | 1.6% |
| Pro (P) | 8 | 6.0% |
| Ser (S) | 7 | 3.8% |
| Thr (T) | 13 | 3.8% |
| Trp (W) | 2 | 0.5% |
| Tyr (Y) | 6 | 2.2% |
| Val (V) | 12 | 8.7% |
| Pyl (O) | 0 | 0.0% |
| Sec (U) | 0 | 0.0% |
Evolution of PrrA
Amino Acid Conservation Scores
The following scores assist in examining evolutionary relatedness of the amino acid sequence of PrrA to that of other amino acid sequences.
- POS: The position of the AA in the SEQRES derived sequence.
- SEQ: The SEQRES derived sequence in one letter code.
- 3LATOM: The ATOM derived sequence in three letter code, including the AA's positions as they appear in the PDB file and the chain identifier.
- SCORE: The normalized conservation scores.
- COLOR: The color scale representing the conservation scores (9 - conserved, 1 - variable).
- CONFIDENCE INTERVAL: When using the bayesian method for calculating rates, a confidence interval is assigned to each of the inferred evolutionary conservation scores.
- CONFIDENCE INTERVAL COLORS: When using the bayesian method for calculating rates. The color scale representing the lower and upper bounds of the confidence interval.
- MSA DATA: The number of aligned sequences having an amino acid (non-gapped) from the overall number of sequences at each position.
- RESIDUE VARIETY: The residues variety at each position of the multiple sequence alignment.
POS SEQ 3LATOM SCORE COLOR CONFIDENCE INTERVAL CONFIDENCE INTERVAL COLORS MSA DATA RESIDUE VARIETY (normalized) 1 D ASP12: -0.486 7 -0.925,-0.243 8,6 9/50 D,N,R 2 R ARG13: 0.275 4 -0.243, 0.523 6,3 13/50 K,P,Q,R,Y 3 S SER14: -0.632 7 -0.925,-0.412 8,6 17/50 K,N,S,T 4 L LEU15: -0.678 7 -0.925,-0.559 8,7 41/50 A,I,L,V 5 L LEU16: -1.003 8 -1.234,-0.813 9,8 41/50 A,F,H,L 6 L LEU17: -0.158 5 -0.412,-0.044 6,5 47/50 I,L,V 7 V VAL18: -1.013 8 -1.134,-0.925 9,8 47/50 A,I,L,V 8 D ASP19: -1.191 9 -1.337,-1.134 9,9 47/50 D,E 9 D ASP20: -1.421 9 -1.455,-1.455 9,9 47/50 D 10 D ASP21: -0.714 7 -0.925,-0.559 8,7 47/50 D,E,N 11 E GLU22: 2.102 1 2.258, 2.258 1,1 47/50 A,D,E,I,K,N,P,Q,R,S,T,V 12 P PRO23: 0.507 3 0.201, 1.009 4,2 47/50 A,D,E,G,K,L,N,P,S 13 F PHE24: -0.580 7 -0.813,-0.412 8,6 47/50 F,H,I,L,T,V 14 L LEU25: -0.433 6 -0.692,-0.243 7,6 47/50 A,C,G,L,R,S,V 15 K LYS26: 1.480 1 1.009, 2.258 2,1 47/50 D,E,G,H,K,N,Q,R,T,W 16 R ARG27: 1.029 2 0.523, 1.009 3,2 47/50 A,F,G,I,L,M,R,S,T,V,W,Y 17 L LEU28: -0.541 7 -0.813,-0.412 8,6 47/50 F,I,L,M,T,V 18 A ALA29: 0.933 2 0.523, 1.009 3,2 47/50 A,E,G,I,N,Q,R,S,T 19 K LYS30: 1.705 1 1.009, 2.258 2,1 47/50 A,F,G,I,K,L,Q,R,T,V,Y 20 A ALA31: -0.159 6 -0.412,-0.044 6,5 47/50 A,C,G,I,L,M,N,R,S,T 21 M MET32: -0.961 8 -1.134,-0.813 9,8 47/50 A,F,I,L,M 22 E GLU33: 0.254 4 -0.044, 0.523 5,3 47/50 A,E,G,K,Q,R,S,T,V 23 K LYS34: 2.007 1 2.258, 2.258 1,1 47/50 A,D,E,F,G,K,L,M,Q,R,S,T 24 R ARG35: 0.749 3 0.201, 1.009 4,2 47/50 A,D,E,L,M,N,Q,R,S,W 25 G GLY36: -1.275 9 -1.455,-1.234 9,9 47/50 G,K 26 F PHE37: 0.226 4 -0.044, 0.523 5,3 47/50 F,H,L,M,Y 27 V VAL38: 1.276 1 0.523, 2.258 3,1 47/50 A,D,E,K,L,N,Q,R,S,T,V 28 L LEU39: -0.651 7 -0.813,-0.412 8,6 47/50 A,C,L,P,T,V 29 E GLU40: 1.033 2 0.523, 1.009 3,2 47/50 A,D,E,F,H,I,K,L,R,S,T,V,Y 30 T THR41: 1.772 1 1.009, 2.258 2,1 47/50 A,C,E,H,I,L,M,Q,S,T,V,W 31 A ALA42: -0.654 7 -0.813,-0.559 8,7 47/50 A,F,H,T,V,Y 32 Q GLN43: 2.160 1 2.258, 2.258 1,1 47/50 A,D,E,F,G,H,N,Q,R,S,T,Y 33 S SER44: -0.228 6 -0.412,-0.044 6,5 47/50 D,G,H,L,N,S,T 34 V VAL45: -0.378 6 -0.692,-0.243 7,6 46/50 A,G,L,S,T,V 35 A ALA46: 2.116 1 2.258, 2.258 1,1 47/50 A,D,E,H,K,L,N,P,Q,R,S,T,V 36 E GLU47: 1.145 1 0.523, 2.258 3,1 47/50 A,D,E,G,M,N,Q,R,S,T 37 G GLY48: -0.725 7 -0.925,-0.559 8,7 47/50 A,F,G,L,V 38 K LYS49: 0.885 2 0.523, 1.009 3,2 47/50 D,E,H,I,K,L,M,R,V,W,Y 39 A ALA50: 2.079 1 2.258, 2.258 1,1 47/50 A,D,E,H,K,L,Q,R,T 40 I ILE51: 2.142 1 2.258, 2.258 1,1 47/50 A,D,E,F,G,I,K,L,M,Q,S,V,W 41 A ALA52: 1.233 1 0.523, 2.258 3,1 47/50 A,F,G,I,L,M,V 42 Q GLN53: 1.647 1 1.009, 2.258 2,1 47/50 A,E,H,I,K,L,M,N,P,Q,R,S,T,V 43 A ALA54: 0.986 2 0.523, 1.009 3,2 47/50 A,D,E,I,N,P,R,S,T 44 R ARG55: 1.930 1 2.258, 2.258 1,1 50/50 A,D,E,G,I,K,L,N,Q,R,V,Y 45 P PRO56: 2.213 1 2.258, 2.258 1,1 50/50 A,D,E,F,H,I,K,M,P,Q,R,T,V 46 P PRO57: 0.463 4 0.201, 1.009 4,2 50/50 A,F,G,N,P,R,V,Y 47 A ALA58: -0.631 7 -0.813,-0.412 8,6 50/50 A,D,G,H,K,N,Q,S 48 Y TYR59: -0.004 5 -0.243, 0.201 6,4 50/50 A,C,H,I,L,P,V,Y 49 A ALA60: -0.030 5 -0.243, 0.201 6,4 50/50 A,C,I,L,V 50 V VAL61: 0.210 4 -0.044, 0.523 5,3 50/50 F,I,L,V 51 V VAL62: -0.346 6 -0.559,-0.044 7,5 50/50 C,F,I,L,M,S,T,V 52 D ASP63: -1.423 9 -1.455,-1.455 9,9 50/50 D 53 L LEU64: -0.362 6 -0.559,-0.243 7,6 50/50 I,L,M,V,W 54 R ARG65: -0.682 7 -0.925,-0.559 8,7 50/50 G,K,M,N,R,T,V,W 55 L LEU66: -0.789 7 -0.925,-0.692 8,7 50/50 L,M 56 E GLU67: -0.694 7 -0.925,-0.559 8,7 50/50 A,D,E,G,P,S 57 D ASP68: 0.334 4 -0.044, 0.523 5,3 50/50 D,E,G,K,N,R 58 G GLY69: 0.102 5 -0.243, 0.523 6,3 50/50 D,E,G,I,K,L,M,T,V 59 N ASN70: -0.540 7 -0.692,-0.412 7,6 50/50 D,N,S,T 60 G GLY71: -1.297 9 -1.455,-1.234 9,9 50/50 A,G 61 L LEU72: -0.217 6 -0.559,-0.044 7,5 50/50 F,I,L,M,V,W 62 D ASP73: -0.002 5 -0.243, 0.201 6,4 50/50 A,D,E,G,H,K,Q,S 63 V VAL74: -0.427 6 -0.692,-0.243 7,6 50/50 A,F,I,L,T,V 64 V VAL75: 0.158 4 -0.243, 0.523 6,3 50/50 A,C,F,I,L,V 65 E GLU76: 0.207 4 -0.044, 0.523 5,3 50/50 A,D,E,H,K,P,Q,R,Y 66 V VAL77: 1.376 1 1.009, 2.258 2,1 50/50 A,D,E,I,K,M,N,Q,R,T,V,Y 67 L LEU78: -0.552 7 -0.813,-0.412 8,6 50/50 I,L,M,V,W 68 R ARG79: -0.539 7 -0.692,-0.412 7,6 50/50 G,K,L,N,Q,R 69 E GLU80: 1.449 1 1.009, 2.258 2,1 50/50 A,D,E,G,I,K,Q,R,S,V 70 R ARG81: 1.916 1 1.009, 2.258 2,1 50/50 A,D,E,H,I,K,L,M,N,Q,R,S,T,V,W 71 R ARG82: 1.282 1 1.009, 2.258 2,1 50/50 A,D,E,G,H,K,N,Q,R,S,Y 72 P PRO83: 0.133 5 -0.243, 0.523 6,3 45/50 A,D,I,K,L,P,Q,T,V,Y 73 D ASP84: 0.789 3 0.523, 1.009 3,2 50/50 A,D,E,G,H,L,M,N,Q,R,T,W 74 C CYS85: 0.574 3 0.201, 1.009 4,2 50/50 A,C,F,I,L,M,T,V 75 R ARG86: -0.924 8 -1.134,-0.813 9,8 50/50 A,K,P,R 76 I ILE87: -0.659 7 -0.813,-0.559 8,7 50/50 I,M,S,V 77 V VAL88: -0.565 7 -0.813,-0.412 8,6 50/50 A,I,L,M,V 78 V VAL89: -0.368 6 -0.559,-0.243 7,6 50/50 F,I,L,M,V 79 L LEU90: -0.765 7 -0.925,-0.559 8,7 50/50 I,L,M,V 80 T THR91: -1.242 9 -1.337,-1.134 9,9 50/50 S,T 81 G GLY92: -1.171 9 -1.337,-1.032 9,8 50/50 A,G,S 82 Y TYR93: -0.715 7 -0.925,-0.559 8,7 50/50 F,H,K,L,Q,R,S,Y 83 G GLY94: -0.342 6 -0.559,-0.044 7,5 50/50 A,D,G,N,S 84 A ALA95: 0.022 5 -0.243, 0.201 6,4 50/50 A,D,E,N,S,T 85 I ILE96: -0.472 6 -0.692,-0.243 7,6 50/50 E,F,I,L,V 86 A ALA97: 0.457 4 0.201, 1.009 4,2 50/50 A,D,E,F,I,M,P,S,T 87 T THR98: -0.835 8 -1.032,-0.692 8,7 50/50 A,D,E,H,L,M,N,T 88 A ALA99: -1.253 9 -1.337,-1.134 9,9 50/50 A,E,K,R 89 V VAL100: -1.099 8 -1.234,-1.032 9,8 50/50 I,L,V 90 A ALA101: 0.796 2 0.523, 1.009 3,2 50/50 A,D,E,F,I,K,L,N,Q,S 91 A ALA102: -1.232 9 -1.337,-1.134 9,9 50/50 A,G,T 92 V VAL103: -0.327 6 -0.559,-0.044 7,5 50/50 A,F,I,L,M,S,T,V,Y 93 K LYS104: -0.119 5 -0.412, 0.201 6,4 50/50 A,D,E,K,N,Q,R 94 I ILE105: 2.072 1 2.258, 2.258 1,1 50/50 A,C,D,G,H,I,K,L,M,Q,R,S,T,V 95 G GLY106: -1.382 9 -1.455,-1.337 9,9 50/50 G 96 A ALA107: -1.353 9 -1.455,-1.337 9,9 50/50 A,S,V 97 T THR108: -0.134 5 -0.412, 0.201 6,4 50/50 A,D,F,L,Q,T,V,Y 98 D ASP109: -1.303 9 -1.455,-1.234 9,9 50/50 D,E,S 99 Y TYR110: -0.709 7 -0.925,-0.559 8,7 50/50 F,Y 100 L LEU111: -0.714 7 -0.925,-0.559 8,7 50/50 I,L,M,V 101 S SER112: -0.199 6 -0.412,-0.044 6,5 50/50 A,C,E,I,P,S,T,V 102 K LYS113: -1.412 9 -1.455,-1.337 9,9 50/50 K 103 P PRO114: -1.394 9 -1.455,-1.337 9,9 50/50 P 104 A ALA115: -0.767 7 -1.032,-0.559 8,7 42/50 A,C,F,L,V 105 D ASP116: -0.564 7 -0.813,-0.412 8,6 42/50 A,D,E,G,H,N,S 106 A ALA117: 0.457 4 -0.044, 1.009 5,2 41/50 A,D,F,I,K,L,N,P,T 107 N ASN118: -0.387 6 -0.692,-0.243 7,6 41/50 A,D,E,G,K,N,R,T 108 E GLU119: -0.108 5 -0.412, 0.201 6,4 41/50 A,D,E,K,N,Q,R,V 109 V VAL120: -0.464 6 -0.692,-0.243 7,6 38/50 I,L,M,T,V 110 T THR121: 1.560 1 1.009, 2.258 2,1 23/50 A,D,I,L,N,Q,R,T,V,Y 111 H HIS122: -0.539 7 -0.813,-0.243 8,6 23/50 A,E,H,L,N,Q 112 A ALA123: -0.970 8 -1.134,-0.813 9,8 23/50 A,L,R,S,T 113 L LEU124: -0.153 5 -0.559, 0.201 7,4 19/50 I,L,V 114 L LEU125: 0.411 4 -0.044, 1.009 5,2 15/50 E,H,I,K,L,Q,R 115 A ALA126: -0.472 6 -0.813,-0.243 8,6 15/50 A,Q,R,S,T 116 K LYS127: 0.296 4 -0.412, 0.523 6,3 11/50 A,I,K,L,N,R,V 117 G GLY128: -0.161 6 -0.692, 0.201 7,4 8/50 G,L,S,V 118 E GLU129: -0.964 8 -1.234,-0.813 9,8 8/50 E,R 119 S SER130: -0.336 6 -0.813,-0.044 8,5 7/50 A,H,R,S 120 L LEU131: -0.401 6 -0.925,-0.044 8,5 7/50 G,L,S 121 P PRO132: 0.446 4* -0.243, 1.009 6,2 6/50 E,K,N,P
=Phylogenetic Tree of PrrA=ш [1]
1umq
Reference
1. Eraso, J. M., J. H. Roh, X. Zeng, S. J. Callister, M. S. Lipton and S. Kaplan. 2008. Role of Global Transcriptional Regulator PrrA in Rhodobacter sphaeroides 2.4.1: Combined Transcriptome and Proteome Analysis. Journal of Bacteriology 190: 4831-4848.
2. Eraso, J. M. and S. Kaplan. 1994. prrA, a Putative Response Regulator Involved in Oxygen Regulation of Photosynthesis Gene Expression in "Rhodobacter sphaeroides". Journal of Bacteriology 176:32-43.
3. Laguri, C., M. K. Phillips-Jones, and M. P. Williamson. 2003. Solution Structure and DNA Binding of the Effector Domain from the Global Regulator PrrA (RegA) from "Rhodobacter sphaeroides": Insights into DNA Binding Specificity. Nucleic Acids Research 31:6778-6787.
4. Zeilstra-Ryalls, J. H. and S. Kaplan. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cellular and Molecular Life Sciences 61:417-436.
- Page Edited by Yana Fedotova, email: yana.fedotova@gmail.com, yfedot@bgsu.edu
- Acknowledgment to Dr. Jill Zeilstra-Ryalls, Susana Retamal and Adam Meade