This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
X-ray crystallography
From Proteopedia
(→See Also - adding content) |
(→See Also - polishing) |
||
| Line 14: | Line 14: | ||
*[http://en.wikipedia.org/wiki/X-ray_crystallography X-ray Crystallography at Wikipedia] | *[http://en.wikipedia.org/wiki/X-ray_crystallography X-ray Crystallography at Wikipedia] | ||
*[http://hamptonresearch.com/gallery.aspx Protein Crystal Gallery] | *[http://hamptonresearch.com/gallery.aspx Protein Crystal Gallery] | ||
| + | |||
*[[Quality assessment for molecular models]] | *[[Quality assessment for molecular models]] | ||
Revision as of 00:24, 19 May 2009
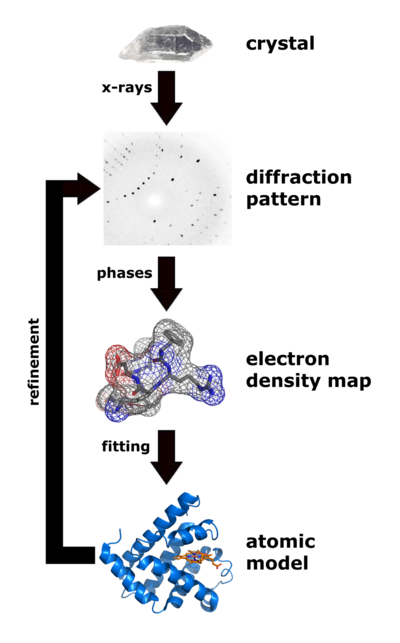
|
| Flow chart showing the major steps in X-ray protein crystallography. (Image from Wikimedia courtesy Thomas Splettstoesser. |
About 85% of the models (entries) in the World Wide Protein Data Bank were determined by X-ray crystallography. (Most of the remaining 15% were determined by solution nuclear magnetic resonance.) Protein crystallography remains very difficult, despite many recent advances. For every new protein sequence targeted for X-ray crystallography, about one in twenty is solved[1][2]. Publication of solved structures involves depositing an atomic coordinate file (PDB file) in the World Wide Protein Data Bank.
See Also
Further Reading
- Crystallography Made Crystal Clear: a guide for users of macromolecular models, a book by Gale Rhodes.
