User:Ramiro Barrantes/FpgNeiRepair
From Proteopedia
(Difference between revisions)
(→Functional Units) |
(→Functional Units) |
||
| Line 27: | Line 27: | ||
| <scene name='User:Ramiro_Barrantes/Workbench/Stabilityintercalationloop/1' target="fpg">D110,F108,R113,R112,F114</scene> | | <scene name='User:Ramiro_Barrantes/Workbench/Stabilityintercalationloop/1' target="fpg">D110,F108,R113,R112,F114</scene> | ||
| <scene name='User:Ramiro_Barrantes/Workbench/Econeiintercalationloopstabili/1' target="ecoNei">Different</scene> | | <scene name='User:Ramiro_Barrantes/Workbench/Econeiintercalationloopstabili/1' target="ecoNei">Different</scene> | ||
| + | | Y || Y || Y || N || N || N || N || N || N || Y | ||
| + | |- | ||
| + | | Intercalation loop | ||
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Stabilityintercalationloop/1' target="fpg">D110,F108,R113,R112,F114</scene> | ||
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Econeiintercalationloop/1' target="ecoNei">Gln76,Met77,Tyr78</scene> | ||
| Y || Y || Y || N || N || N || N || N || N || Y | | Y || Y || Y || N || N || N || N || N || N || Y | ||
|} | |} | ||
| Line 40: | Line 45: | ||
=== Functional Cluster 2: Stability of catalytic helix === | === Functional Cluster 2: Stability of catalytic helix === | ||
[[Image:F2WebLogo.jpg|thumb|166px|left|The LER triad interact in stabilize the catalytic helix (Pro2 and Glu3), it is not clear how this is achieved in the other subfamilies]] The triad Leu4, Glu8 and Arg57 interact and provide stability to helixA, which has the catalytic residue Pro2,Glu3 and Glu6. This triad is present in the same four clades as above (Fpg1, Fpg2 and Plants and Fungi). This triad is not present in the remaining clades and it is not clear how the same stability is provided. Leu211 also has a hydrophobic interaction with Leu4. | [[Image:F2WebLogo.jpg|thumb|166px|left|The LER triad interact in stabilize the catalytic helix (Pro2 and Glu3), it is not clear how this is achieved in the other subfamilies]] The triad Leu4, Glu8 and Arg57 interact and provide stability to helixA, which has the catalytic residue Pro2,Glu3 and Glu6. This triad is present in the same four clades as above (Fpg1, Fpg2 and Plants and Fungi). This triad is not present in the remaining clades and it is not clear how the same stability is provided. Leu211 also has a hydrophobic interaction with Leu4. | ||
| - | === Functional Cluster 4: Intercalation Loop === | + | === Functional Cluster 4: Intercalation Loop === |
| - | + | [[Image:IntercalationLoop.jpg|thumb|166px|left|The residue in positions 77 and 78 suggest a possible intercalation loop] | |
| + | The intercalation loop inserts into the space left by the excised base. | ||
</div> | </div> | ||
Revision as of 17:54, 21 May 2009
Contents |
The FpgNei Protein Superfamily
Functional Units
| G. Stereothermophilus Fpg | E. Coli Nei | ||||||||||||
|
|
| Functional Cluster | Variant 1 | Variant 2 | Fpg1 | Fpg2 | Plant | Neil1 | Neil2 | Neil3 | Proteo | Actino1 | Actino2 | MimiVirus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Support for perfectly conserved Asn168 | Y | Y | N | N | N | N | N | N | N | N | ||
| Stability of catalytic helix | Y | Y | Y | N | N | N | N | N | N | N | ||
| Stability of intercalation loop | Y | Y | Y | N | N | N | N | N | N | Y | ||
| Intercalation loop | Y | Y | Y | N | N | N | N | N | N | Y |
Functional Cluster 1: Stability of perfectly conserved Asn168
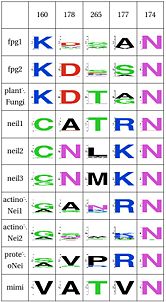
Asn74, along with two other amino acids have an effect in the orientation and kinking of the DNA. In 4 of the 9 clades (Fpg1, Fpg2 and Plants and Fungi) Asn174 is supported by Lys160, which in turn hydrogen bonds with Leu249 and Ser250. In the other clades (Actinobacteria 1 and 2, Proteacteria and all vertebrate sequences), Arg171 that comes from a different helix fulfills the same roles as Lys160. One important difference is that the Zinc Finger is shaped differently in the absence of DNA, and there is a hydrogen bond between one of the beta-sheets and the arginine. One hypothesis is that the arginine or the lysine is necessary to support the Asn174, crucial for orientation of the DNA.
Functional Cluster 3: Stability of intercalation loop
Image:IntercalationLoopSupport.jpg
This structure provides stability for the amino acids that insert into the space vacated by the damaged base
Functional Cluster 2: Stability of catalytic helix
The triad Leu4, Glu8 and Arg57 interact and provide stability to helixA, which has the catalytic residue Pro2,Glu3 and Glu6. This triad is present in the same four clades as above (Fpg1, Fpg2 and Plants and Fungi). This triad is not present in the remaining clades and it is not clear how the same stability is provided. Leu211 also has a hydrophobic interaction with Leu4.
Functional Cluster 4: Intercalation Loop
[[Image:IntercalationLoop.jpg|thumb|166px|left|The residue in positions 77 and 78 suggest a possible intercalation loop] The intercalation loop inserts into the space left by the excised base.