Fpg Nei Protein Family
From Proteopedia
Ramiro Barrantes (Talk | contribs)
(New page: {{STRUCTURE_3cin | PDB=1R2Y | SCENE= }} ==DNA Repair and Base Excision DNA Repair== For a video of DNA repair [http://www.youtube.com/watch?v=CcTayxEblio click here]. The picture bel...)
Next diff →
Revision as of 21:36, 18 June 2009
| |||||||||
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
DNA Repair and Base Excision DNA Repair
For a video of DNA repair click here. The picture below also illustrates DNA repair:
 The genome of any living organisms is being continuously affected by exogenous and endogenous agents, such as ultraviolet light, ionizing radiation, different chemicals and the cell's own metabolites (such as reactive oxygen). Therefore, different systems have evolved to repair these damages. With some of these systems shared throughout all lifeforms. Therefore, the proper functioning of DNA repair is critical for survival. There are six pathways of DNA repair (reviewed in Friedberg et al), and one of the is base-excision repair. The latter's distinguishing feature is that it removes lesions as single bases, as opposed to dNMPs or short oligonucleotides like other systems. [1]
The genome of any living organisms is being continuously affected by exogenous and endogenous agents, such as ultraviolet light, ionizing radiation, different chemicals and the cell's own metabolites (such as reactive oxygen). Therefore, different systems have evolved to repair these damages. With some of these systems shared throughout all lifeforms. Therefore, the proper functioning of DNA repair is critical for survival. There are six pathways of DNA repair (reviewed in Friedberg et al), and one of the is base-excision repair. The latter's distinguishing feature is that it removes lesions as single bases, as opposed to dNMPs or short oligonucleotides like other systems. [1]
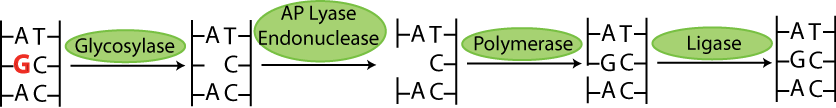
Base excision repair's signature enzyme are the DNA glycosylases. These enzymes work by recognizing and removing a single damaged base from DNA. They are called DNA glycosylases because they hydrolize the N-glycosidic bond of the damaged deoxynucleoside. The subsequent steps of the pathway (strand incision, gap-filling and ligation) are done by other enzymes. See below for a cartoon of the process [2][3]:
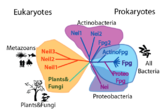
Evolution and related structures
Homologous structures have been solved, including Fpg protein from Lactococcus Lactis (1pjj)[4], Bacillus Stereothermophilus (1r2y)[5], Thermos Thermophilus (1ee8)[6] and Escherichia Coli(1k82)[7] and Nei from Escherichia Coli (1k3w)[8]. The overall structure is similar, and some of the damages include 8-oxoguanine and fapyG (1xc8)[9].