User:Tilman Schirmer/Sandbox 201
From Proteopedia
(→Intro) |
(→Substrate binding) |
||
| Line 29: | Line 29: | ||
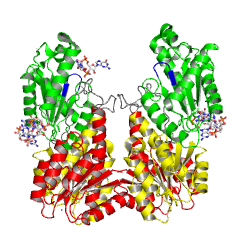
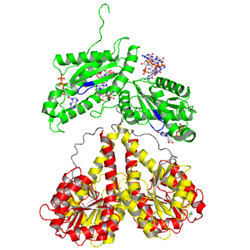
| - | The motif is part of the <scene name='User:Tilman_Schirmer/Sandbox_201/Substrate_binding_site/ | + | The motif is part of the <scene name='User:Tilman_Schirmer/Sandbox_201/Substrate_binding_site/5'>substrate binding site</scene> as identified in the structure of PleD in complex with <scene name='User:Tilman_Schirmer/Sandbox_201/Gtp-a-s/2'>GTP-alpha-S / Mg++</scene>. The GGDEFY domain binds only '''one''' GTP subsrate molecule. For the reaction to proceed, '''two''' GTP loaded GGDEF domains have to align antiparallely. MODEL. |
Revision as of 11:10, 21 June 2009
PleD
Contents |
Overview
|
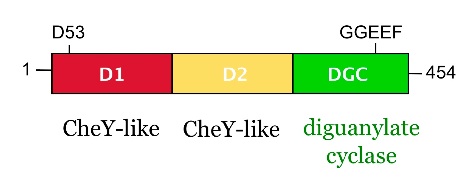
from Caulobacter crescentus is a response regulator with an unorthodox catalytic, diguanylate cyclase, output domain. It is composed of a canonical CheY-like response regulator receiver () domain,
a Rec-like () adaptor domain,
and a C-terminal domain that confers the catalytic acitvity.
The GGDEF domain is named after the highly conserved (in PleD it is GGEEF) that locates to a β-hairpin.
Substrate binding
|
The motif is part of the as identified in the structure of PleD in complex with . The GGDEFY domain binds only one GTP subsrate molecule. For the reaction to proceed, two GTP loaded GGDEF domains have to align antiparallely. MODEL.
Allosteric product binding site
|
C-di-GMP
Primary inhibition site (Ip)
Secondary inhibition site (Is)
Primary and secondary inhibition sites
Two conformations
|
|