Green Fluorescent Protein
From Proteopedia
(→Notes and Literature References) |
|||
| Line 41: | Line 41: | ||
==Notes and Literature References== | ==Notes and Literature References== | ||
<references/> | <references/> | ||
| - | + | ||
==Additional Literature and Resources== | ==Additional Literature and Resources== | ||
*Roger Y. Tsien (1998) The Green Fluorescent Protein. Annual Review of Biochemistry 67, 509-544. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/9759496 9759496] | *Roger Y. Tsien (1998) The Green Fluorescent Protein. Annual Review of Biochemistry 67, 509-544. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/9759496 9759496] | ||
Revision as of 10:14, 13 July 2009
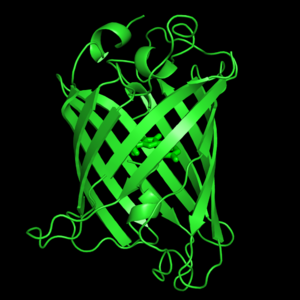
Green Flourscent Protein
with flourescence-conferring chromophore
at center of the barrel-like structure
Contents |
Background
Osamu Shimomura, Martin Chalfie and Roger Y. Tsien shared the 2008 Nobel Prize in Chemistry for their for the discovery and development of the green fluorescent protein, GFP.
GFP is small protein (21 kDa) that naturally occurs in the jellyfish Aequorea victoria and does not require cofactors to become fluorescent. The chromophore at the center of the structure that is responsible for its fluorescence is formed
spontaneously from a tri-peptide motif in the primary structure of GFP. Being small and facile has lead to its use by investigators as a tool to examine a multitude of processes in many organisms; often in such research, GFP is fused to other proteins genetically.
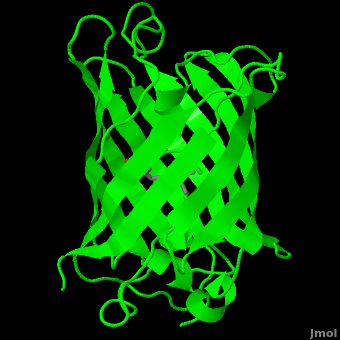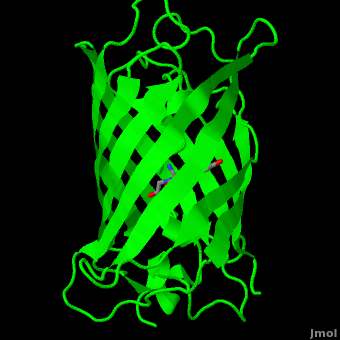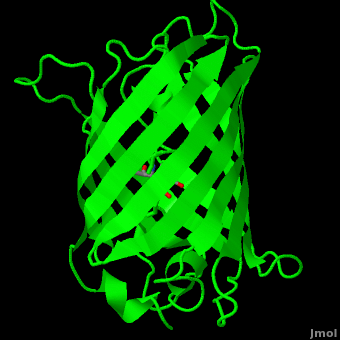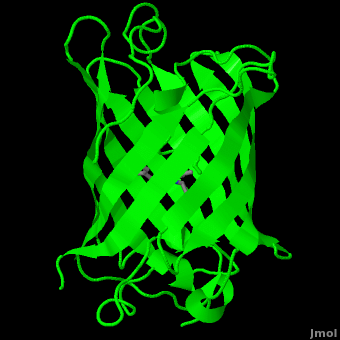
Structure
| |||||||
The crystal structure of GFP [1][2] is an eleven-stranded anti-parallel beta-barrel, threaded by an alpha-helix, running up along the axis of the cylinder. In the structure is colored by secondary structure(Alpha Helices and Beta Strands ) to better show the eleven strands and the helices.
:
The chromophore is in the distorted alpha-helix that runs along the axis of the can, close to the center of the can-like cylinder.
The chromophore is formed from the tripeptide motif Serine65-Tyrosine66-Glycine67 after translation and folding of GFP. As the GFP protein folds into its native conformation, these three amino acids are forced into a sharp turn, greatly favoring a nucleophilic attack of the amide of Glycine67 on the carbonyl of Serine65, leading to imidazolinone formation by cyclization and dehydration. At this point, GFP is not fluorescent; however, in the presence of molecular oxygen, the α–β bond of Tyrosine66 is subsequently dehydrogenated into conjugation with the imidazolinone, which results in maturation of the GFP chromophore to its fluorescent form [3][4].
The chromophore .
Reference for the Structure
Crystal structure of the Aequorea victoria green fluorescent protein., Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ, Science. 1996 Sep 6;273(5280):1392-5. PMID:8703075
Related Structures and Topics
- 1ema Aequorea victoria Green Fluorescent Protein
- 1gfl Aequorea victoria Green Fluorescent Protein
- 1b9c Aequorea victoria Green Fluorescent Protein Mutant F99s, M153t And V163a
- GFP featured at the Molecule of the Month series of tutorials by David Goodsell.
Notes and Literature References
- ↑ Crystal structure of the Aequorea victoria green fluorescent protein., Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ, Science 1996 6;273(5280):1392-1395. PMID:8703075
- ↑ The molecular structure of green fluorescent protein., Yang F, Moss LG, Phillips GN Jr, Nat Biotechnol. 1996 Oct;14(10):1246-51. PMID:9631087
- ↑ Wavelength mutations and posttranslational autoxidation of green fluorescent protein., Heim R, Prasher DC, Tsien RY, Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12501-4. PMID:7809066
- ↑ Understanding, improving and using green fluorescent proteins. Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY, Trends Biochem Sci. 1995 Nov;20(11):448-55.PMID:8578587
Additional Literature and Resources
- Roger Y. Tsien (1998) The Green Fluorescent Protein. Annual Review of Biochemistry 67, 509-544. PMID:9759496
- GFP featured at the Molecule of the Month series of tutorials by David Goodsell.
- The GFP site by Marc Zimmer, Ph. D., at Connecticut College, who authored Glowing Genes: A Revolution In Biotechnology.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Wayne Decatur, Karsten Theis, Eran Hodis, Laura Carbone, Karl Oberholser, Mark Hoelzer, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Marius Mihasan, Joseph M. Steinberger, David Canner