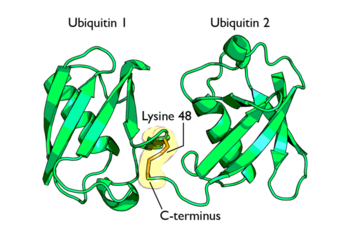This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Ubiquitin Structure & Function
From Proteopedia
Ubiquitin is a single 8565 Mr polypeptide consisting of 76 amino acid residues. Ubiquitin is highly known for its role in ATP-dependant protein degradation.
| |||||||||
| 1ubq, resolution 1.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Introduction
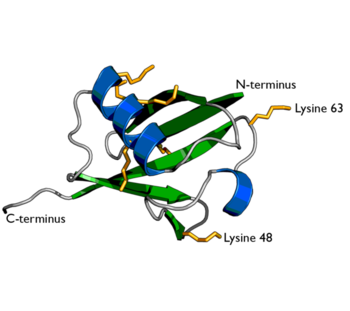
Ubiquitin is one of the most highly conserved eukaryotic proteins. Primary structures found throughout ubiquitin are identical in all bovine, insects and human ubiquitin. The only difference observed amongst these species is seen in the terminal Gly-Gly residues. Yeast and oat ubiquitin only differ in three of the 76 residues when compared to ubiquitin found in higher eukaryotes. Ubiquitin can not only be found in the nucleus, but in the cytoplasm and cell-surface membrane as well. One interesting characteristic of ubiquitin is its stability. Ubiquitin is able to withstand a range of pH levels and temperatures and is very resistant to tryptic digestion, while still containing seven Lysine and four arginine residues. Many aspects of ubiquitin's structure aid in this durability. Ubiquitin contains a hydrophobic core. Three hydrophobic residues found on the α-helix and 11 of the 13 hydrophobic residues from the β-sheet are involved in constructing this hydrophobic core. The main contributor to the ubiquitin stability is the vast amount of hydrogen-bonding interactions observed. The whole structure of ubiquitin undergoes significant hydrogen bonding, aside from the COOH terminus.
Function
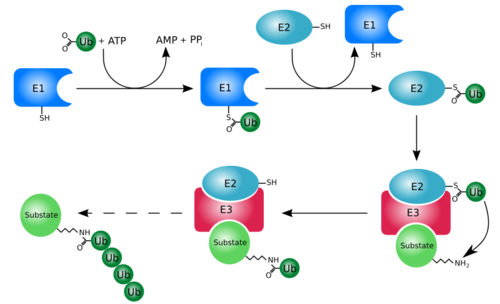
At first, ubiquitin was believed to be a hormone involed in inducing the differentiation of lymphocytes and activating adenylate cyclase [1]. However, today, ubiquitin is primarily known for its role in intracellular ATP-dependent protein degradation. This is accomplished through the process of several seperate reactions:
Activation
The first step of ubiquitin activation involves the formation of a ubiqiotin-adenylate intermediate. This reaction requires an E1 ubiquitin-activating enzyme. The second step of ubiquitin activation transfers ubiquitin to the E1 active site cysteine residue and AMP is released. This step results in a thioester linkage between the C-terminal carboxyl group of ubiquitin and the E1 cysteine sulfhydryl group.
Ubiquitin Conjugation
The activated ubiquitin in then transferred to a ubiquitin-conjugating enzyme, E2 through a trans-thiolesteration reaction. Ubiquitin is then transferred to the ε-amino group of a lysine chain on the target protein. An isopeptide bond is now formed between the carboxyl terminal of ubiquitin and the ε-amino group of the target protein lysine residue. This is accomplished by E2 directly.
Some proteins may be selected for degradation through the use of protein E3. E3 binds and catalyzes the isopeptide bond between ubiquitin and the target protein. Several activated ubiquitin may be added while still bound to E2 following the first ubiquitin addition.
Proofreading
Conjugate Metabolism
- ↑ Vijay-Kumar, S., Begg, CE., Wilkinson, KD and Cook, WJ. 1985. Three-dimensional structure of ubiquitin of 2.8 angstrom resolution. Proc Natl Acad Sci 82:3582-3585
Proteopedia Page Contributors and Editors (what is this?)
Jaclyn Gordon, Joel L. Sussman, Michal Harel, David Canner, Andrea Gorrell, Alexander Berchansky, Karsten Theis