Copper, Zinc Superoxide Dismutase
From Proteopedia
Copper, zinc superoxide dismutase is an oxidoreductase enzyme responsible for the very rapid two-step dismutation of the toxic superoxide radical to molecular oxygen and hydrogen peroxide through alternate reduction and oxidation of the active-site copper[1].
Contents |
Introduction
Oxygen is vital to sustain life; our cells cannot live without it. Oxygen is the final electron acceptor in the electron transport chain, allowing us to produce a large amount of energy from our food. But oxygen is also a dangerous compound.[2]. Reactive forms of oxygen, such as superoxide, leak from the respiratory chain and wreak havoc on the cell. Superoxide is a free radical, a molecule that readily accepts electrons, which make them highly reactive. They can strip electrons from proteins, lipids, or nucleic acids, thereby destroying their function and possibly resulting in cell death.
Copper, zinc superoxide dismutase is an important antioxidant defense in nearly all cells exposed to oxygen. It forms a crucial component of the cellular response to oxidative stress by detoxifying the superoxide radical via a special reaction known as dismutation [3].
Superoxide Dismutase Family
In mammals there are three known isomers of superoxide dismutase (SOD). Copper and Zinc containing SOD1 is located in the cytoplasm. Manganese containing SOD2 is located in the mitochondria while a second Copper and Zinc containing SOD3 is located in the extracellular space. SOD3 and SOD2 are tetramers whereas SOD1 is a dimer. These enzymes perform the dismutation reaction by a similar mechanism.
General Structure
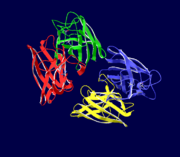
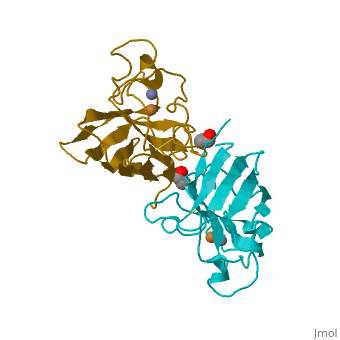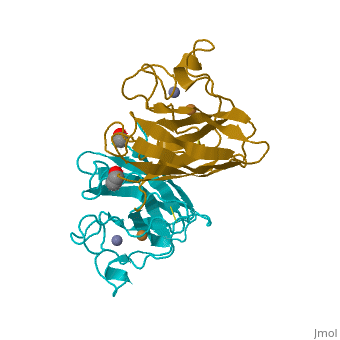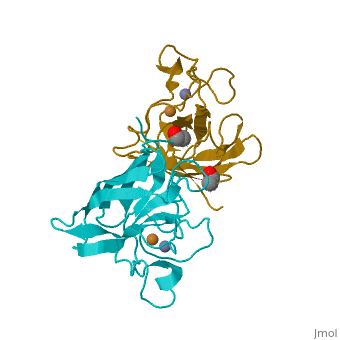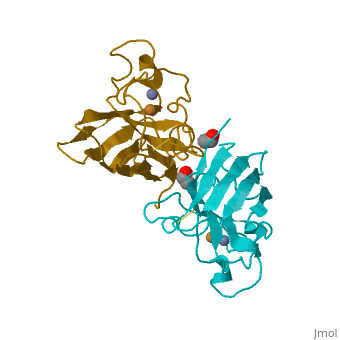
Copper, zinc superoxide dismutase is a metalloprotein, containing both copper and zinc ions in each subunit, where copper acts as the active site. It is a homotetramer, containing four polypeptide chains (B,O,G,Y), 152 residues in length. Each chain contains one alpha-helix and 12-14 beta-sheets. It has a structural weight of 62913.70 with the ligand identifier of C2 H4 O.
Each enzyme subunit is composed primarily of eight antiparallel β strands that form a flattened cylinder, plus three external loops. The largest loop includes both a disulfide region and a Zn-liganding region, each of which resembles one of the other two loops in overall structure. The second largest loop includes a short section of α helix. The smallest loop forms a Greek key connection across one end of the β barrel. The single disulfide bond, which forms a left-handed spiral, covalently joins the largest loop to the beginning of β strand 8[4].
Oxidative Capabilities
Free radicals such as superoxide are a type of reactive oxygen species that can strip electrons from proteins, lipids, or nucleic acids, thereby destroying their function and resulting in cell dysfunction or death. They are highly reactive molecules with an unpaiared electron in an outer orbital, which can initiate chain reactions by removal of an electron from another molecule to complete its own orbital[5].
Apparently, if SOD is defective, superoxide is not degraded and can destroy cells. Motor neurons appear to be particularly sensitive to superoxide attack. Mutations in human CuZnSOD are associated with the development of familial amyotrophic lateral sclerosis (motor neuron disease)[3]
The Reaction of Dismutase
|
Dismutation is a term that refers to a special type of reaction, where two equal but opposite reactions occur on two separate molecules [2].
Superoxide is produced both accidentally and also as a reactive oxygen species required for a number of enzyme-catalyzed reactions. Copper, zinc superoxide dismutase catalyzes the reaction between superoxide and water to yield oxygen and hydrogen peroxide. Most enzymes that produce and require superoxide are in the peroxisomes,together together with superoxide dismutase, catalase and peroxidases[6]. Copper, zinc superoxide dismutase takes two molecules of superoxide, strips the extra electron off of one, and places it on the other. So, one ends up with an electron less, forming normal oxygen, and the other ends up with an extra electron. The one with the extra electron then rapidly picks up two hydrogen ions to form hydrogen peroxide.
![]()
However, hydrogen peroxide is also dangerous and needs to be detoxified. The cells use the enzyme catalase, a ubiquitous heme protein that catalyzes the dismutationof hydrogen peroxide into water and molecular oxygen.
2H2O2 → 2H2O + O2
Importance of Vitamins
Free radicals are molecules that can react and alter the structure of molecules that control the normal cell function of the cell. According to the free radical theory, this damage can accumulate with age, leading to the normals symptoms seen with aging. Antioxidants, such as beta carotene, vitamin C, and vitamin E, can donate electrons to free radicals, without themselves becoming harmful. Thus, antioxidants may prevent the damage caused by free radicals and may ward off age-related disorders, ranging from wrinkles to cancer. Experiments designed to test this hypothesis, however, have not consistently produced positive results[7]
One of the long-term benefits of exercise may be to increase the amount of superoxide dismutase in the cell. The elevated aerobic metabolism during exercise causes more ROS to be generated. In response, the cell synthesizes more protective enzymes. The net effect is one of protection, because the increase in superoxide dismutase more effectively protects the cell during periods of rest [8].
See Also
Free-Radical Theory of Aging (FRTA)
References
- ↑ Tainer JA, Getzoff ED, Richardson JS, Richardson DC. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983 Nov 17-23;306(5940):284-7. PMID:6316150
- ↑ 2.0 2.1 http://www.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/pdb94_1.html
- ↑ 3.0 3.1 Hough MA, Hasnain SS. Structure of fully reduced bovine copper zinc superoxide dismutase at 1.15 A. Structure. 2003 Aug;11(8):937-46. PMID:12906825
- ↑ Tainer JA, Getzoff ED, Beem KM, Richardson JS, Richardson DC. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J Mol Biol. 1982 Sep 15;160(2):181-217. PMID:7175933
- ↑ Metabolic Pathways and their Control pg.574
- ↑ Free Radicals and Antioxidant Nutrients ch.6 pg.484
- ↑ Seeley, Stephens, Tate
- ↑ Oxidative Phosphorylation Ch.18, pg. 518
Proteopedia Page Contributors and Editors (what is this?)
Jordan Schibli, Jane S. Richardson, David Canner, Michal Harel, Andrea Gorrell