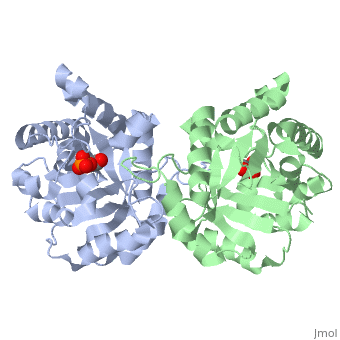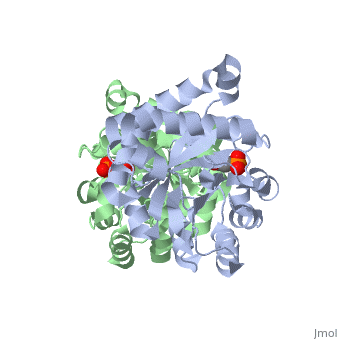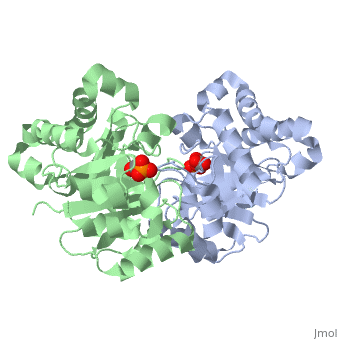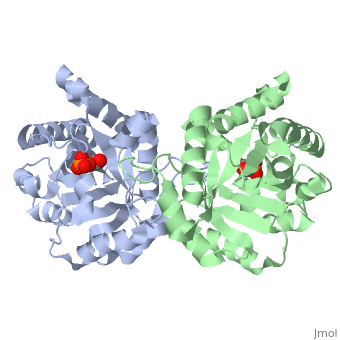2ypi
From Proteopedia
|
CRYSTALLOGRAPHIC ANALYSIS OF THE COMPLEX BETWEEN TRIOSEPHOSPHATE ISOMERASE AND 2-PHOSPHOGLYCOLATE AT 2.5-ANGSTROMS RESOLUTION. IMPLICATIONS FOR CATALYSIS
Overview
The binding of the transition-state analogue 2-phosphoglycolate to triosephosphate isomerase from yeast has been investigated crystallographically. An atomic model of the enzyme-inhibitor complex has been refined against data to 2.5-A resolution to a final R factor of 0.18. The interactions between the inhibitor and enzyme have been analyzed. The inhibitor forms hydrogen bonds to the side chains of His 95 and Glu 165. The latter hydrogen bond confirms that Glu 165 is protonated upon PGA binding. The structure of the complexed enzyme has been compared to that of the unbound form of the enzyme, and conformational changes have been observed: the side chain of Glu 165 moves over 2 A and a 10-residue flexible loop moves over 7 A to close over the active site. Spectroscopic results of phosphoglycolic acid binding to triosephosphate isomerase that have been amassed over the years are also explained in structural terms. The implications for catalysis are noted.
About this Structure
2YPI is a Single protein structure of sequence from Saccharomyces cerevisiae with as ligand. The following page contains interesting information on the relation of 2YPI with [The Glycolytic Enzymes]. Active as Triose-phosphate isomerase, with EC number 5.3.1.1 Full crystallographic information is available from OCA.
Reference
Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-A resolution: implications for catalysis., Lolis E, Petsko GA, Biochemistry. 1990 Jul 17;29(28):6619-25. PMID:2204418
Page seeded by OCA on Thu Feb 21 18:57:07 2008