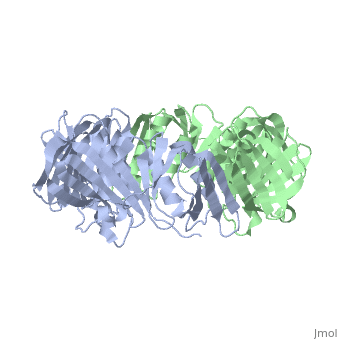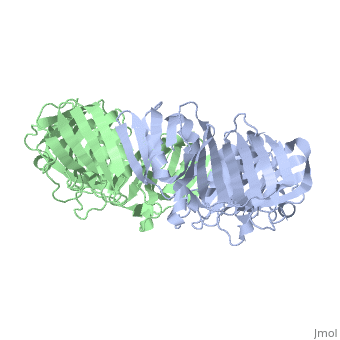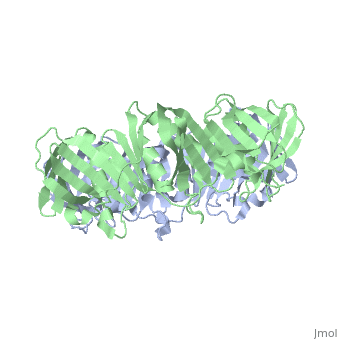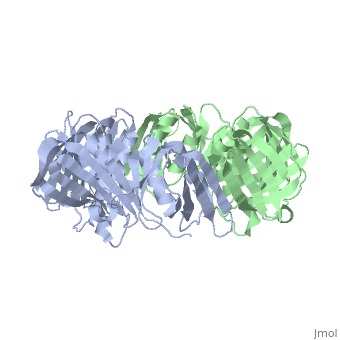
Structure
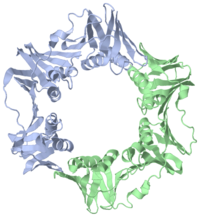
The beta subunit binds DNA by forming a ring around the DNA helix, essentially acting as a sliding clamp, also known as a beta clamp. This conformation allows the enzyme to move along the DNA structure without diffusing away, thereby increasing the processivity and rate of nucleotide polymerization. The beta complex consists of a dimer of monomers that form a donut-type structure with an ~80A total diameter. This creates an inner ring of approximately 35A, in which A- and B-DNAs have room to bind. The dimeric ring is a pseudo-symmetrical six pointed star, with each monomer made up of three structurally similar domains. These domains align head to tail to form the ring shape.
Function
Model studies of B-DNA binding this enzyme suggest that the beta subunit is specifically designed to limit DNA associations in order to move freely along the helix, ensuring high processivity. This decreased association with DNA is due to the fact that the protein alpha helices do not actually enter the major and minor grooves of the DNA in favor of simply spanning over them. The beta clamp in the Pol III holoenzyme must be reloaded onto the template strand of DNA at a rate of once per second, in order to achieve the observed rate of 1,000 nucleotides per second observed in E. Coli DNA. This reloading is done by the gamma subunit of the holoenzyme, also known as the clamp loader. After the beta subunit is bound to the DNA, the core of the holoenzyme dissociated from the clamp loader to allow for DNA replication. When Pol III encounters the previously synthesized Okazaki fragment, the beta subunit dissociates from the DNA and is ready to bind subsequent templates.
About this Structure
2pol is a 2 chain structure with sequence from Escherichia coli. The June 2012 RCSB PDB Molecule of the Month feature on Sliding Clamps by David Goodsell is 10.2210/rcsb_pdb/mom_2012_6. Full crystallographic information is available from OCA.