Sandbox Reserved 390
From Proteopedia
Human topoisomerase II beta in complex with DNA and etoposide
| |||||||||||
References
- ↑ Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011 Jul 22;333(6041):459-62. PMID:21778401 doi:10.1126/science.1204117
- ↑ Kathryn L. Gilroy, Chrysoula Leontiou, Kay Padget, Jeremy H. Lakey and Caroline A. Austin* "mAMSA resistant human topoisomerase IIβ mutation G465D has reduced ATP hydrolysis activity” Oxford JournalsLife Sciences Nucleic Acids Research Volume 34, Issue 5Pp. 1597-1607. DOI: 10.1093/nar/gkl057
- ↑ Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011 Jul 22;333(6041):459-62. PMID: 21778401./>. This structure provided the first observation of a TOP2 ternary cleavage complex by an anticancer drug.
The high-resolution structure of the hTOP2βcore-DNA-etoposide ternary complex reveals the intricate interplays between , and . This aspect is extremely important because all vertebrates possess two highly similar yet functionally distinct TOP2 isoforms. The α-isoform is particularly important for DNA replication and is usually present at high levels in fast growing cancer cells, whereas the β-isoform is mainly involved in transcription related processes. Although the inhibition of both TOP2 isoforms contributes to the drug-induced death of cancer cells, targeting of the β-isoform has been implicated in deleterious therapy related secondary malignancies. Therefore, it is desirable to develop the isoform specific TOP2-targeting agents.
COMPOUND ACTIVE SITE
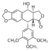
This molecule has 36 represented with magenta helices and 40 represented with blue arrows in the . At the active site we can see how the (in green) is stabilized. The detail view of the show how the ligand is stabilized by amino acid (at position 503B in blue), (at position 479B in yellow with hydrogen bond interaction), (at position 778B in purple), also by nucleoside (in light blue at position 9F) and (in green at possition 13D showing a Pi-Pi interaction)
ETOPOSIDE RESISTANCE
The interplay between the protein, the DNA, and the drug explains the structure-activity relations of etoposide derivatives and the molecular basis of drug-resistant mutations. This resistance occurs via two mechanisms: 1) Decreased accumulation via increased P-glycoprotein ; and 2) Changes in target proteins (mutation or decreased expression of topoisomerase II or decreased apoptosis due to mutation of P53).
1. Decreased accumulation via increased P-glycoprotein (a multidrug resistance): This drug resistance mechanism is characterized by decreased intracellular accumulation of drug facilitated by overexpression of the human multidrug resistance (mdrl) gene, causing overproduction of P-glycoprotein. This cell membrane protein acts as an export pump for a wide variety of unrelated foreign natural products. By maintaining lower intracellular levels of drug, lower drug concentration would be available to the target, which is topoisomerase II.
[[Image:PGP.gif|thumb|right|500 px|P-glycoprotein as a transmembrane drug efflux pump<ref>doi:10.1038/nrc823</li></ol></ref>