CBI Molecule Workshop
From Proteopedia
Contents |
Create Your Own Proteopedia Page
Use the right Internet browser
Proteopedia and Jmol requires Java for 64-bit browsers. Therefore, use Firefox (works best for page making) or Safari, and not Chrome.
Create a page
- At the 'Search' box on the left of any proteopedia page, type your page title. For a draft page for this class, type:
Your Username/Sandbox1
You will be asked to click on a red linked caption to create your new page.
Edit the page
Create all your content in the 'edit this page' tab. Remember that you need to log in to edit a page. Changes are recorded and can be undone in the 'history' tab, just like Wikipages.
Create written content
Typography
Simple and useful basics...
- In the 'edit this page' tab, write something, then select your sentence or word. Click the appropriate buttons to create Bold or italics fonts, or "Headers".
- Click the button "Save page" at the page bottom, and you'll see how it will look like on your Proteopedia page.
Tips:
You can combine those typography: eg. bold+italic, bold+italic+header.
When you create headers, Proteopedia automatically generates a content box at the top of the page to contain all the headers of the page.
To create more complicated constructions of your proteopedia page, browse other proteopedia pages, and observe the syntax used to create the special effects.
Link or reference other webpages
The 'External link' button lets you create a link to another webpage (other than Proteopedia pages). Example: Research articles related to your page.
- Click the 'External link' button. You'll see: [http://www.example.com link title]. Paste in your web address of interest. When you create a link title, it will show up as a blue linked caption like this: UMASS Amherst
The 'Internal link' button lets you create a link to another Proteopedia webpage. Example: a Proteopedia page that contains a PDB molecule related to your page, your Proteopedia profile page.
- Click the 'Internal link'. You'll see: [[Link title]]. Paste in the Proteopedia page of interest. It will show up as a blue linked caption like this: CBI Molecules
Create still images
The 'Embedded image' button lets you include images into your page. Example: gene pathways, chemical reactions, still structural images from Pymol.
- Ensure that you are logged on, because images are uploaded to your user account.
- Click the 'Upload file' tab on the left of the page. Upload an image of interest. Copy the file name of the image.
- When in the 'edit this page tab', click the 'Embedded image' button and paste the file name into [[Image:Example.jpg]].
- You can change the size and position of the image by including further details such as: [[Image:Example.jpg||300px|left|]]
Create dynamic structural content
Structure that appears on your page front
- Click the '3D' button. You will see:
<Structure load='Insert PDB code or filename here' size='500' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' />
For this workshop:
- Type '7DFR' in place of 'Insert PDB code or filename here'.
- Type 'DHFR is pretty cool' in place of 'Insert caption here'.
You can choose published or your own PDB file later for your own Proteopedia page.
- Click the button "Save page" at the page bottom, and you'll see how it will look like on your Proteopedia page.
Creating green-linked scenes Scenes are specific dynamic display of your structure of interest. Try these steps to get a taste of scene-creating:
- In the 'edit this page' tab, click the 'show' button of
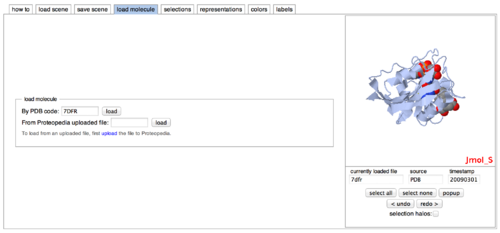
- Load a molecular structure onto your page by first clicking the 'load molecule' tab. For this workshop, type '7DFR'. Click 'load'. You'll see the DHFR protein molecule in the box on the right, as such:
- Click 'selections' tab. For this workshop, click 'helix' and 'add to selection'. In the box on the right, check mark 'selection halos' to see which part of the protein is selected. Selection will be highlighted in yellow.
- Click 'representations' tab. Check mark 'meshribbon' and click 'set representation'. You'll see that the yellow highlighted sections change to mesh representation.
- Click 'colors' tab. When 'selection' is selected, click a green color box. When 'background' is selected, click a black color box.
- Click 'labels' tab. When 'atom' is selected, click 'select none' button in the right hand side box. Click an atom on the protein, so that only one residue is selected. In the text box, type "Just playing around". Click 'set label'.
- Click 'save scene' tab. Type 'Scene 1'. Click 'Save current scene'. You will see a Wikitext:
Copy and paste the following line where you want the scene link to appear (scroll down if needed) and edit the TextToBeDisplayed:
- Copy the scene name in full, i.e.: <scene name='56/565760/Scene_1/2'>TextToBeDisplayed</scene>
- In the main editing box, paste the scene name in full.
- Within '<scene name='56/565760/Scene_1/2'>TextToBeDisplayed</scene>' type 'Playing around with DHFR' in place of "TextToBeDisplayed". This text will show up as the caption of your green link.
- Click 'hide' in the Scene authoring tools.
- Click the 'Save page' button to view your output.
- To display this molecule in your page, click the 'save scene' tab.
- Type into the 'Scene name' box, for example: Scene 1
- Click 'Save current scene'. A wikitext will appear as:
Copy and paste the following line where you want the scene link to appear (scroll down if needed) and edit the TextToBeDisplayed: <scene name='56/565760/Scene_1/1'>TextToBeDisplayed</scene>
- Select and copy the line '<scene name='56/565760/Scene_1/1'>TextToBeDisplayed</scene>'
- In the main editing box of the 'edit this page' tab, paste '<scene name='56/565760/Scene_1/1'>TextToBeDisplayed</scene>'. 'TextToBeDisplayed' is where you can type a caption name that will turn into a green link that is used to initiate visualization of the molecule.
- Save page to view your output.
== Extra fun stuff: Display evolutionary conservation ==
Go to the ConSurf Server and follow the instructions to generate a conservation score of your protein. Extract the RasMol Coloring Script.
Consur http://consurf.tau.ac.il/
Favorite tips
Observe other molecules
Creating a great page is a reiterative process!
Remember to give credit to images appropriately, and avoid plagiarism like the plague by writing in your own words.
Your Heading Here (maybe something like 'Structure')
| |||||||||||