Sandbox Reserved 764
From Proteopedia
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
|
Contents |
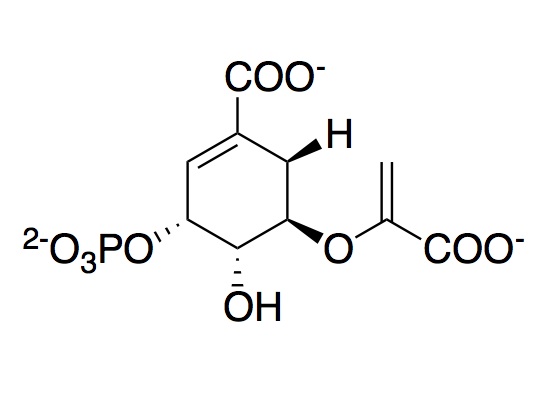
5-Enolpyruvylshikimate-3-phosphate synthase
Formula weight: 47244.52 Da
Classification: Transferase
Length: 427 residues
Domains: 2
The monomeric enzyme 5-Enolpyruvlyshikimate-3-phosphate synthase (EPSP synthase) is an enzyme involved in the shikimate pathway found only in plants and microorganisms. The shikimate pathway is essential for the biosynthesis of chorismate, a precursor to majority of the aromatic compounds produced in the cell, including the aromatic amino acids (L-tyrosine, L-phenylalanine, L-tryptophan),that are needed for protein synthesis. Studies suggest that the compounds produced in this pathway constitute as much as or more than 35% of the dry mass of plants.
EPSP Synthase Structure
- add monomer, weight, number off domains, residues within each domain**
The three-dimensional structure of EPSP synthase in Escherichia coli was determined using X-ray crystallography. The enzyme has two distinct domains, each of which have a radius of approximately 25 Angstroms. The two domains are linked by two crossover chain segments with both the amino and carboxyl terminus of the polypeptide chain located in the lower domain. The domains ppear to be formed by a 6-fold replication of a protein folding unit consisting of two parallel helices and a four-stranded sheet. Each domain is formed from three of these units, forming a threefold symmetry axis.
EPSP Synthase Mechanism
EPSP synthase catalyzes the reversible transfer of the enolpyruvyl group from phosphoenolpyruvate (PEP) to shikimate-3-phosphate (S3P) to form the products EPSP and inorganic phosphate. Steady-state and pre-steady-state kinetic studies indicate that the kinetically preferred reactions pathway occurs with S3P binding to EPSP synthase. PEP preferentially interacts with the E-S3P complex. Chemical modification studies on EPSPS indicate that Lys, Arg, and His residues are essential for activity of the enzyme. The reactive Lysine and Arginine residues have been identified as Lys-23 and Arg-28. Mutation of Lys-23 to Arg does not affect the activity of the enzyme. However, mutation of Lys-23 to either Ala or Glu leads to complete loss of enzyme activity. It is speculated that both Lys-23 and Arg-28 constitute a part of the active site recognizing the anionic residues on the substrates. During the EPSP synthase reaction, the C-O bond of the peptide is cleaved.
Implicatons
The implications in studying EPSP synthase involve the fact that this enzyme is not present in humans but is essential for bacteria and apicomplexan parasites. This makes EPSP synthase a selective target for antimicrobial drugs and decreases possible negative impacts of drugs in humans.