Heterotrimeric G-proteins family
Human heterotrimeric G-proteins are derived from 35 genes: 16 encoding α subunits, 5 β and 14 γ subunits.
The α subunits function as guanine nucleotide on-off switches, mechanistically similar to other G-proteins that are enzymatic GTPases.
G-proteins interact with diverse protein partners, such as G-protein coupled receptors (GPCRs), downstream effectors, and other proteins.
One important G-protein interaction is with members of the RGS protein family. This interaction occurs when the G-protein alpha subunit is activated, and depends on the Gα class, which in turn depends on their sequence that classifies them into several sub-types. Based on the Phylogenetic tree of mammalian G-protein, G-protein α-subunits classified to 4 groups based on there sequence identity. The first two groups, Gαi and Gαq families have selectivity towards The RGS members of R4-subfamily and R12-subfamily. another group, Gαz subunits were suggested to have selectivity for the RGS members of RZ-subfamily. The last subfamilies Gαs and Gα12/13 might doing interaction with diverse proteins subfamilies that include the ~120-residue RGS homology domain but can't interact with canonical RGS members.
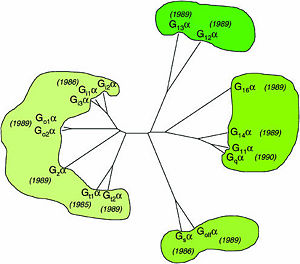

Phylogenetic tree of mammalian G-protein α-subunits classified to 4 groups based on there sequence identity.[1]
RGS proteins
Regulator of G-proteins signaling (RGS) proteins play a critical role in many G protein-dependent signaling pathways. Thus, RGS proteins have been implicated in a wide range of pathologies, including cancer, hypertension, arrhythmias, drug abuse and schizophrenia. RGS proteins ‘turn off’ heterotrimeric (αβγ) G-proteins and thereby determine the duration of G protein–mediated signaling events. Therefore, RGS proteins function as GTPase Activating Proteins (GAPs), and this GAP activity is mediated by allosteric interactions.
RGS proteins are selective for binding to the transition state of Gα(GTP → GDP + Pi).
Like many signaling proteins, RGS proteins comprise a large and diverse family. In human genome, Thirty-seven RGS proteins are encoded by gene loci; this collection of related proteins has been divided into 10 different subfamilies based on the relatedness of their RGS domain sequence and their multiple domain architectures. The major subfamily contains About 20 ‘canonical’ RGS proteins that can in theory downregulate any of the 16 activated Gα subunits, although in practice they interact only with members of the Gi and Gq families. In these proteins, the ~120-residue RGS homology domain functions as a GTPase-activating protein (GAP) for GTP-bound Gα subunits. In addition to these domains, diverse proteins subfamilies include additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity. R7-subfamily members share a multi-domain protein architecture composed of DEP and GGL domains on the N-terminal side of the RGS domain. R12-subfamily members possess a tandem repeat of Ras binding domains (RBDs) and a single GoLoco motif.
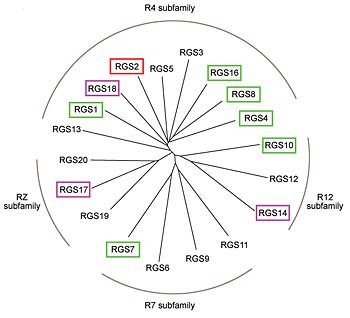

Phylogenetic tree of 19 human RGS domains.
RGS proteins whose activity was tested are colored by their GAP activity, RGS proteins with high GAP activity (green), RGS proteins with low but discernible activities (purple) and RGS2 had no measurable activity (red).
[2]
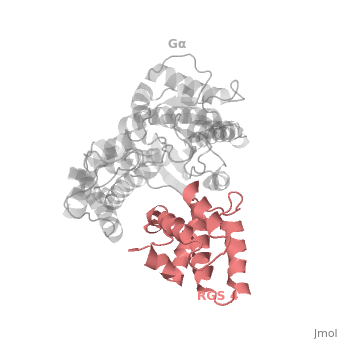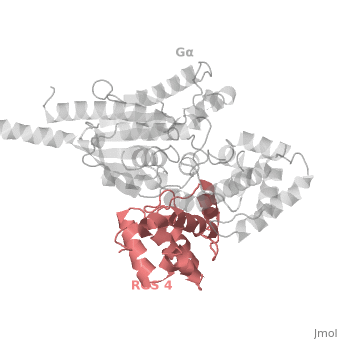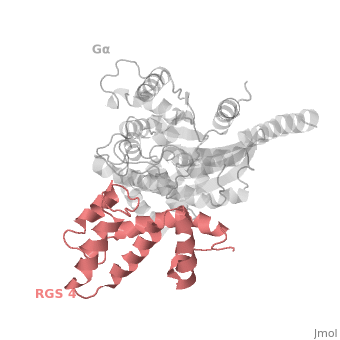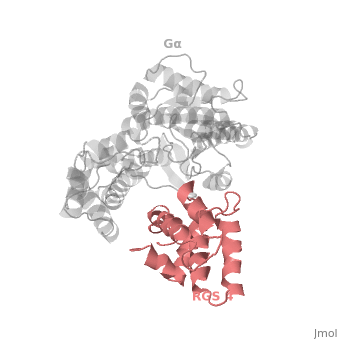
RGS4 Structural highlights
1AGR is a complex of RGS4 and Gαi1 proteins defined by X-ray crystallographic analysis.
corresponds to an array of nine α-helices: α1, α2, α3, α4, α5 and α6 helices are colored in blue, aqua, yellow, coral, magenta and dark green respectively, while helices α7, α8 and α9 are colored red. The 3D structure of RGS4 protein fold into , the terminal subdomain colored magenta, contains the N and C termini of the box and is formed by α1, α2, α3, α8, and α9; and the larger bundle subdomain colored cyan formed by α4, α5, α6, and α7. The both two subdomains are required for RGS4 GAP activity.
Gαi1 Structural highlights
Gαi1 subunits adopt a conserved fold of that composed of six α helices shown as blue cartoon, and a conserved GTPase domain shown in gray cartoon. The GTPase domain hydrolyzes GTP and provides most of Gα's binding surfaces for Gβγ, receptors, effectors and RGS proteins. contains three flexible regions designated switch-I presented as blue sticks, switch-II presented as magenta sticks and switch-III presented as green sticks that change conformation in response to GTP binding and hydrolysis, GDP–Mg+2 bound in the active site of Gαi1 is shown as a ball-and-stick model. The three switch regions of Gαi1 residues: 176–184, 201–215, and 233–241, respectively. [3]
RGS-G proteins interactions
Many RGS protein residues located in the vicinity of the (RGS protein shown as wheat cartoon and Gαi1 subunit shown as white surface) contribute to RGS-G proteins interaction. Based on energy calculation and experimental validation of RGS-Gα complexes from Gαi subfamily members, these residues classified into two major groups: shown as red spheres that are located mainly in the center of the RGS domain–Gα interface and have a primary role in accelerating Gα GTPase by stabilizing Gα in an optimal conformation for GTP hydrolysis. Whereas the shown as purple spheres are located mostly at the periphery of the interface where they contribute to Gα subunit's recognition.[4]
Gα subunits participate in a range of interactions with a variety of other proteins. Therefore, they have interfaces that interact selectively with receptors, effector subfamilies and RGS proteins. However, from Gαi subfamily members that interact specifically with RGS proteins are highly conserved (red spheres). These Gα Residues located on Gα switch regions interact with Significant & Conserved RGS residues because of the pivotal role of the switch regions in GTP hydrolysis that is catalyzed by RGS proteins. On the other hand, Gα residues located in switch regions II and III and multiple residues in the Gα all-helical domain interact with Modulatory RGS residues. For example, one important RGS4 residue that projects into the active site of Gαi is r-Asn-128, which contacts the side chains of a-Gln-204, a-Ser-206, and a-Glu-207.[5]