This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
We start with the primary structure of the molecule. DNA's are quite important in that they determine the identity, and therefore the potential, of any given strand.
Function
Disease
Relevance
Structural highlights
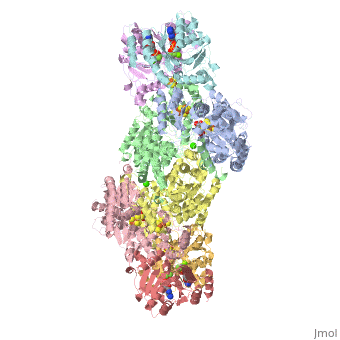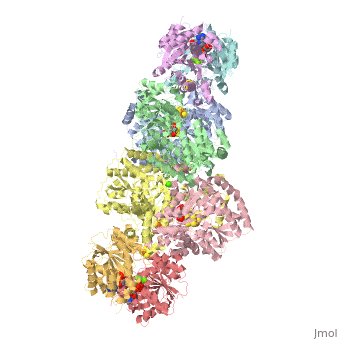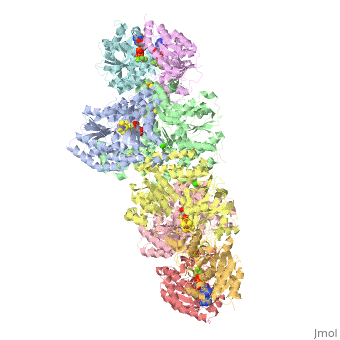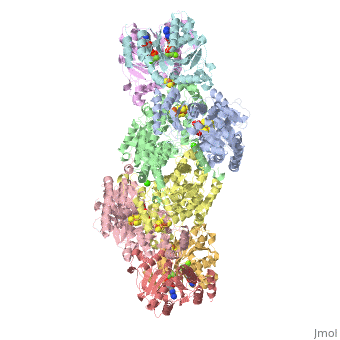
The structure of nitrogenase consists of metal clusters centered throughout the protein. Three of these clusters are the 4Fe-4S Cluster, P-Cluster, and FeMo-Cluster. At either end of the protein are two copies of the Fe protein dimer, which is also the site of the ATP binding site. At these sites are ADP molecules which form a stable complex with the Fe protein. The MoFe protein, the central components of the protein, is where most of nitrogenase’ function is carried out. This protein requires a constant state of electrons which is supplied by the Fe protein which uses the hydrolysis of ATP to pump these electrons into the MoFe protein. Thus, it is helpful for the Fe protein to be coupled with ADP/ATP at the ends of the protein.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT by starting from scratch or loading and editing one of these sample scenes.