This is a default text for your page Steve Klimcak/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
Structure
domains
This is the
Active Site
Function
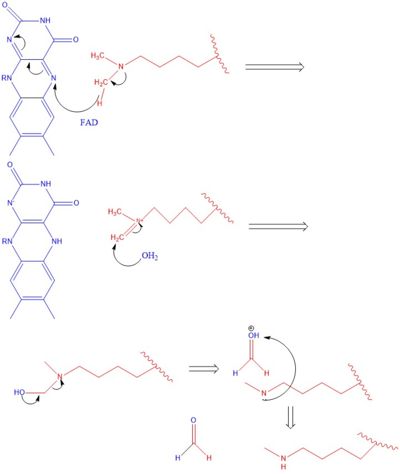
Mechanism
The methylation of lysine via LSD1 occurs through a 2 electron process. Shown in Figure 3 the first step involves the initiating a hydride transfer from the one of the two methyl groups bound to the nitrogen at the lysine tail, via a N5 on the flavin group. An imine cation is then formed at the end of the lysine tail to compensate for the loss of hydride. In the next step the imine is hydrolyzed and transitions to an Hemiaminal. This then breaks down easily to formaldehyde and the product lysine.
Hydrophobic Pocket
The hydrophobic pocket located in the active site cavity of LSD1, forms a catalytic chamber where the substrate lysine is oriented and positioned to interact with the
FAD co-factor to initiate demethylation. The specific residues making up the pocket include valine-317, glycine-330, alanine-331, methionine 332, valine-333, phenylalanine-338, leucine-569, asparagine-660, lysine-661, tryptophan 695, serine 749, serine 760, and tyrosine-761. The shown in green surrounds the FAD in a way such that it exposes the that is responsible for the two electron demethylation. Another three seperate pockets help bind the histone tail residues to the substrate lysine which is essential for identifying different modifications of the histone tail.
Application
Disease
Relevance
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.