User:Clara Costa D'Elia/Sandbox 1
From Proteopedia
Light Harvesting Complex II
This is a default text for your page Clara Costa D'Elia/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
introductionPhotosynthesis represents the main source of energy sustaining life on earth. The primary process of photosynthesis starts with the absorption of sunlight by an arrangement of photosynthetic pigments embedded into a proteic matrix called the light harvesting (LH) antenna complexes. The excitation energy of photosynthetic pigments is then transferred to the photosynthetic reaction center where it is converted into chemical energy. .[3] . Typically, purple bacteria contain two types of antenna complexes, both of which are integral membrane proteins. LHl, is intimately associated with the reaction centre forming the so-called 'core' complex. Arranged more peripherally to this, and present in variable amounts, is the second type, LH2. Both types of complex are built on a similar modular principle. In order to increase the spectral cross-section of absorption, purple bacteria also produce light-harvesting complexes. In most cases a primary light-harvesting complex (LH1) and peripheral light-harvesting complexes (LH2) are synthesised LH2 complexes are produced in variable amounts according to the available light levels, the absorbance range of the particular LH2 (800 and 850, 800 and 820 nm), the temperature, and the bacterial species and strain (Zuber & Brunisholz, 1991). When purple bacteria are grown under anaerobic conditions they incorporate the photosynthetic apparatus described above into invaginated intracytoplasmic phospholipid membranes.[4] Functionlight-harvesting complexes make possible for Purple bacterial to maximize the spectrum of light avaiable to them, modifyng the absorption properties of their chromophores; The energy absorbed is used in the bacteria photochemistry. In the LHC The proteins determine the disposition of the pigments, therefore changing and influencing their absorption spectra. The properties and times scales of energy transfer arise from the relative pigment interaction energies and pigment site energy disorder. These in turn are controlled by factors such as inter-pigment geometries and their interactions with protein and membrane environments. [5] The light-absorbing pigments, bacteriochlorophyll a (Bchl a) and carotenoids, are non-covalently bound to two low-molecular-weight hydrophobic apoproteins, a and p. The native complexes are oligomers of these components. [6] Each individual light-harvesting complex is composed of oligomers of short peptides (α and β) with associated pigments (Hawthornthwaite & Cogdell, 1991). αβ apoproteins with their non-covalently bound carotenoid and bacteriochlorophyll (Bchl ) pigments form the multi-subunit complexes LH1 and LH2 StructureThe differences between the LH1 and LH2 complexes reside in their protein/pigment stoichiometry and modes of oligomerization. Structural studies have shown that LH2 complexes are formed from eight or nine αβ subunit oligomers Structural studies have shown that LH2 complexes are formed from nine ab , organised in a ring of inner a (hollow cylinder of radius 18 A) and outer b-peptides (outer cylinder of radius 34 A). The a-apoprotein contains 53 amino acids, and the p-apoprotein 41.[7] The Bchl a-binding histidines of the a (His 31) and b (His 30) apoproteins face outwards and inwards, respectively,
forming a complete ring of 18 overlapping Bchl a molecules. For
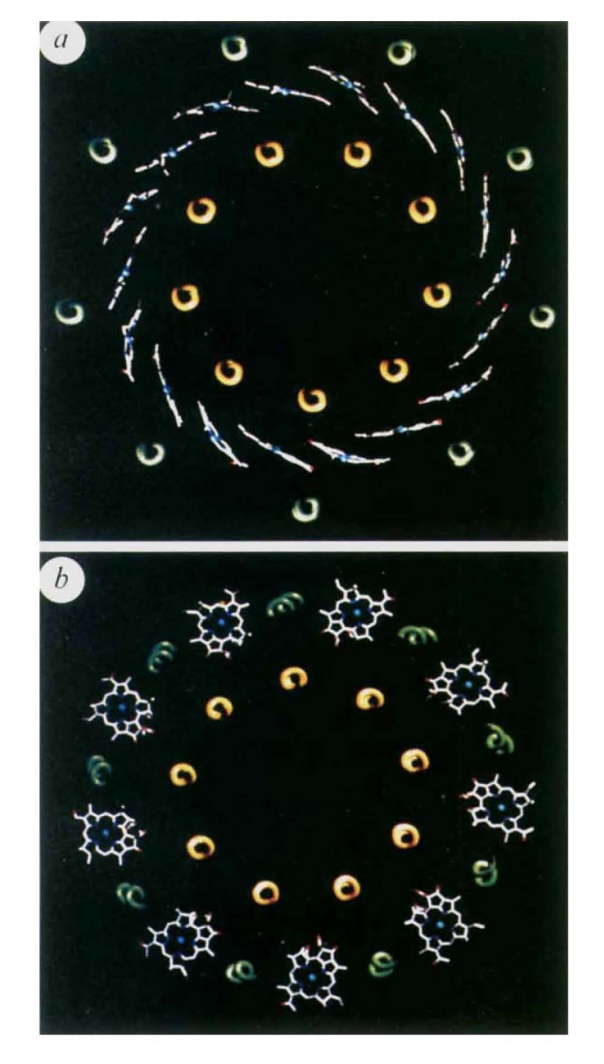
these molecules, the planes of the bacteriochlorins are parallel to
the membrane normal and their centres are approximately loA
from the presumed periplasmic membrane surface. The nine
remaining Bchl a molecules are packed between the p-apoprotein
helices a further 16.5 A into the membrane with their bacteriochlorin rings parallel to the membrane surface.
. Circular dichroism analysis of these absorption bands has led to the conclusion that the Bchl a molecules that absorb at 800 nm are
largely monomeric, whereas those absorbing at 850 nm are
strongly exciton-coupled5
• Conserved histidine residues in the
apoproteins have been shown, by resonance Raman spectroscopy, to be liganded to the Mg at the centre of the Bchl a that
absorbs at 850 nm
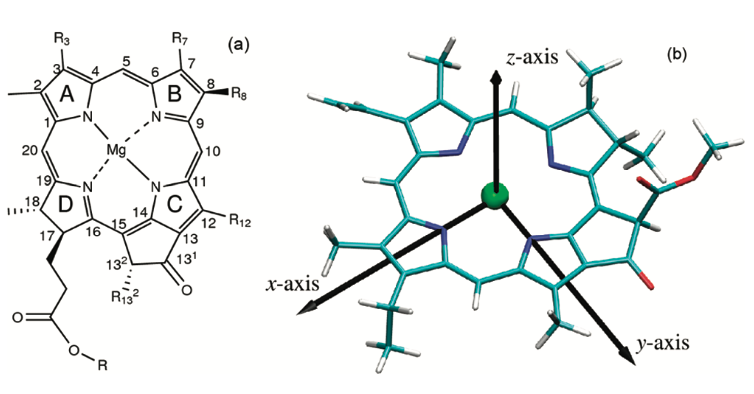
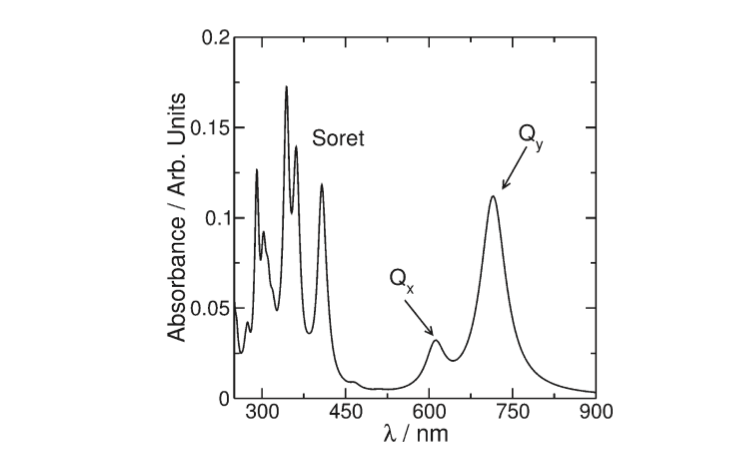
In between the b-peptides and close to the cytoplasmic surface, aswell as Near the periplasmic surface, and between a and b- peptides, are , they are responsible for near infrared absorption, being called B800 and B850. pigments are also present and absorb in the visible part of the spectrum and perform the additional role of protection against photo-induced oxidation, in the case of the LHC II the carotenoid present is rhodopin glucoside. About the pigmentsMost photosynthetics pigments are chlorophylls (Chl), bacteriochlorophylls (BChl), and carotenoids, they represent the keystone for energy storage in photosynthetic organisms The BChl a Qy transition dipole moments are responsible for the near infrared absorption by Bchl a molecules at generic wavelengths 800 nm and 850 nm, and for this reason these pigments are called B800 and B850.[9] Like other dipole moments, the transition dipole refers to a difference in charge from one region of a molecule to another. The transition dipole occurs when an electron is excited from the ground state to an excited state. The charge distribution of the ground state is different from that of the excited state, and it is this change in electron density between the two states that leads to the transitoin dipole. By convention, the y molecular axis of chlorophylls and bacteriochlorophylls is defined as the axis passing through the N atoms of rings A and C; The spectrum of photosynthetic pig- ments exhibits essentially two characteristic absorption bands
(Figure 2): one of them called the Soret band can be found in the
UV region and is a complex band composed of a large series of
electronic transitions. The other called Q is in the visible region
of the spectrum and is the most important for the photophysics
involved in the photosynthetic process
Important to understanding how these complexes trap energy is the degree of delocalisation of the lowest energy states between 850 nm and 870 nm.[13]
The delocalization energy is the extra stability a compound has as a result of having delocalized electrons. Electron delocalization is also called resonance. Therefore, delocalization energy is also called resonance energy. The resonance hybrid is more stable than any of its resonance contributors is predicted to be Charge delocalization is a stabilizing force because it spreads energy over a larger area rather than keeping it confined to a small area. Since electrons are charges, the presence of delocalized electrons brings extra stability to a system compared to a similar system where electrons are localized. The stabilizing effect of charge and electron delocalization is known as resonance energy. Since conjugation brings up electron delocalization, it follows that the more extensive the conjugated system, the more stable the molecule (i.e. the lower its potential energy). If there are positive or negative charges, they also spread out as a result of resonance. Energy transfer mechanismWhen a Bchl molecule is excited by light, its first excited singlet state lasts for a few nanoseconds!. The light-harvesting system must be able to transfer the absorbed energy to the reaction centre in a shorter time than this. Some of the important features that allow this to take place are revealed by the structure reported here. Previous biophysical studies (reviewed in ref. 1) have shown that energy transfer within the LH2 complex can occur from the BSOO to the BS50 BChl a molecules in 0.7 ps. Once the energy reaches the BS50 molecules, it is rapidly transferred among them. This is seen as an ultrafast depolarization of the excited state on the 200-300 fs timescale. Energy transfer from LH2 to LHI occurs in the 5-20 ps time range, but with LH2 alone the decay of the S50 nm excited singlet state takes 1.1 ns. The ring of BS50 Bchl a molecules acts rather like a 'storage ring', with the excited state rapidly delocalized over a large area. The delocalization is facilitated by a highly hydrophobic environment which reduces the dielectric constant, allowing coupling over large distances. The energy is then available for transfer from any part of the ring to any neighbouring LHI complex. It is clear from electron microscopy imaging of the LHI complex 13 , and from a comparison of the primary structures3 of the LH2 and LH I complexes, that the structure of the LHI complex is similar to that of LH2 (with a larger ring). With such a ring structure, there is no requirement for the LHI complex to have a special orientation to receive energy from the LH2 ring. Furthermore, because the BS75 Bchl a molecules in LHI are liganded to homologous histidine residues, as in LH2, it is likely that the BS50 and BS75 bacteriochlorophyll rings will be at the same point in the membrane. The overall effect will be to allow energy transfer from any LH2 to any LHI complex that is within range, without regard to the orientation of either complex. This reduction in the dimensionality of the process will lead to a further kinetic gain. Previous studies have shown that the carotenoid in this LH2 complex acts as an efficient accessory light-harvesting pigment (>50%)7. The excited singlet lifetime of carotenoids is usually less than 10 pS8. Therefore, if energy transfer is to compete successfully with these rapid de-excitation processes, the carotenoid must be located very close to the acceptor bacteriochlorophylls8, as seen in the structure. [15] Structural highlightsThis is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
| ||||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ https://doi.org/10.1021/jp203826q
- ↑ https://doi.org/10.1038/374517a0
- ↑ https://www.sciencedirect.com/science/article/pii/S002228360300024X?via%3Dihub
- ↑ https://doi.org/10.1038/374517a0
- ↑ https://doi.org/10.1038/374517a0
- ↑ https://doi.org/10.1038/374517a0
- ↑ https://doi.org/10.1016/S0022-2836(03)00024-X
- ↑ https://bmc1.utm.utoronto.ca/~vijay/prototype_V12/physChem/molExcit/p08/index.html
- ↑ https://doi.org/10.1021/jp203826q
- ↑ https://doi.org/10.1021/jp203826q
- ↑ https://doi.org/10.1016/S0022-2836(03)00024-X
- ↑ https://chem.libretexts.org/Bookshelves/Physical_and_Theoretical_Chemistry_Textbook_Maps/Supplemental_Modules_(Physical_and_Theoretical_Chemistry)/Chemical_Bonding/Valence_Bond_Theory/Delocalization_of_Electrons
- ↑ https://doi.org/10.1038/374517a0