Sandbox Home
From Proteopedia
table id="tableColumnsMainPage" style="width:100%;border:2px solid #ddd;border-collapse: collapse;table-layout: fixed; "> <tr><td colspan='3' style="background:#F5F5FC;border:1px solid #ddd;">
As life is more than 2D, Proteopedia helps to bridge the gap between 3D structure & function of biomacromolecules
Proteopedia presents this information in a user-friendly way as a collaborative & free 3D-encyclopedia of proteins & other biomolecules.
</td></tr>
<tr> <th style="padding: 10px;background-color: #33ff7b">Selected Research Pages</th> <th style="padding: 10px;background-color: #f1b840">In Journals</th> <th style="padding: 10px;background-color: #79baff">Education</th> </tr>
<tr valign='top'>
<td style="padding: 5px;">HIV-1 protease
by David Canner >>> Visit this page >>> |
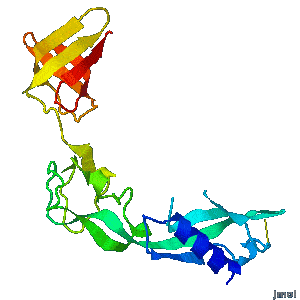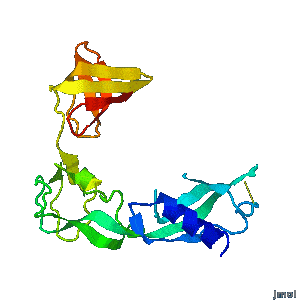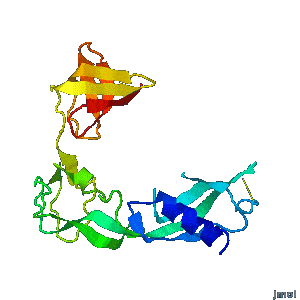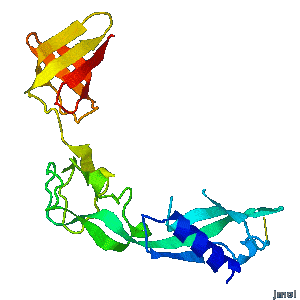
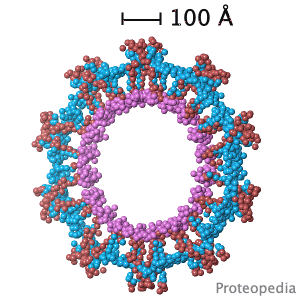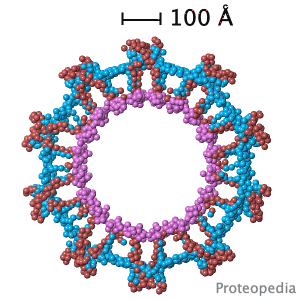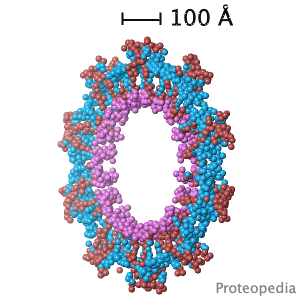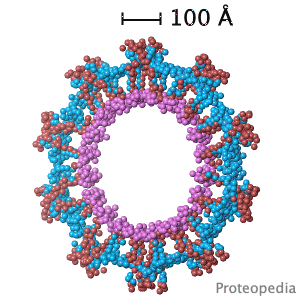
Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica.
H Matsunami, YH Yoon, VA Meshcheryakov, K Namba, FA Samatey. Scientific Reports 2016 doi: 10.1038/srep27399 >>> Visit this I3DC complement >>> |
Eastern Equine Encephalitis virus
Although only a few people in the USA get Eastern Equine Encephalitis every year, the fatality rate is 30%, and many survivors have ongoing neurological problems. The virus is transmitted by mosquitoes from animals, especially birds, to humans. This RNA virus has a complicated capsid (a slab of which is shown) composed of protein shells with an enclosed lipid bilayer. The structures of virus capsids can be explored using free FirstGlance in Jmol. |
</tr>
<tr style="font-size: 1.2em; text-align: center;"> <td style="padding: 10px;background-color: #33ff7b"></td> <td style="padding: 10px;background-color: #f1b840"></td> <td style="padding: 10px;background-color: #79baff"></td> </tr>
<tr style="font-size: 1.0em; text-align: center;">
<td style="padding: 10px;>
How to add content to Proteopedia
Who knows ...
</td>
<td style="padding: 10px;>
About Interactive 3D Complements - I3DCs
How to get an I3DC for your paper
</td>
<td style="padding: 10px;>
Teaching strategies using Proteopedia
Examples of pages for teaching
How to add content to Proteopedia
</td>
</tr>
<tr><td colspan='3' >
| About | Contact | Hot News | Table of Contents | Structure Index | Help |
</td></tr> </table>