Sandbox 22
From Proteopedia
Contents |
Kenny's Sandbox (22)
Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load and display another structure.
| |||||||||
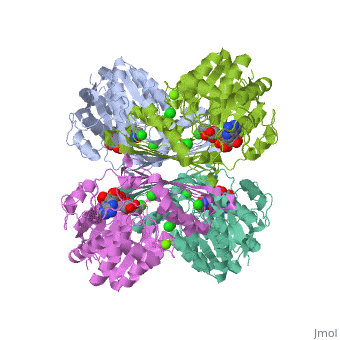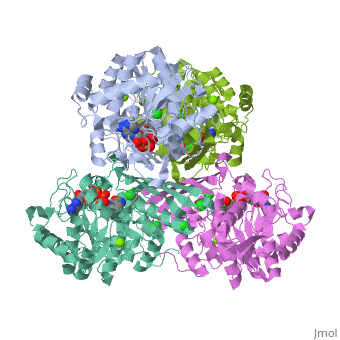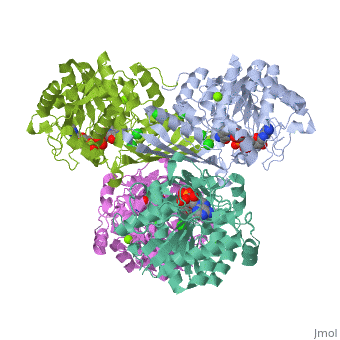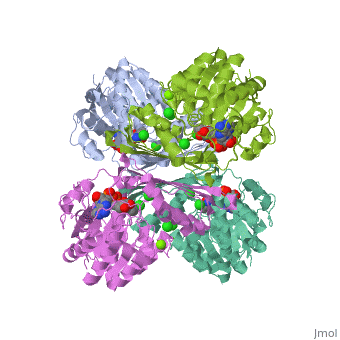
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES
Template:ABSTRACT PUBMED 7651541
About this Structure
2LEF is a 3 chains structure of sequences from Mus musculus. This structure supersedes the now removed PDB entry 1lef. Full experimental information is available from OCA.
Reference
- Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995 Aug 31;376(6543):791-5. PMID:7651541 doi:http://dx.doi.org/10.1038/376791a0
Page seeded by OCA on Tue Feb 17 13:43:41 2009
Categories: Mus musculus | Case, D A. | Li, X. | Love, J J. | Wright, P E. | Dna bending | Dna binding | Hmg | Lef1 | Tcr-a | Transcription factor