User:Wayne Decatur/Haloarcula Large Ribosomal Subunit
From Proteopedia
The Haloarcula Large Ribosomal Subunit
The molecular machine catalyzing petide bond synthesis is a ribozyme.
Introduction
The ribosome is a complex composed of RNA and protein that adds up to several million daltons in size and plays a critical role in the process of decoding the genetic information stored in the genome into protein as outlined in what is now known as the Central Dogma of Molecular Biology. Specifically, the ribosome carries out the process of translation, decoding the genetic information encoded in messenger RNA, one amino acid at a time, into newly synthesized polypeptide chains. The ribosome functions as a complex of two complexes of many proteins and RNAs of substantial length; these two complexes are the small ribosomal subunit and the large ribosomal subunit.
In 2000, the large ribosomal subunit from Haloracula marismortui was solved. Haloracula is a [Extremophiles|halophilic]] archaea.
Thomas Steitz shares a 2009 Nobel Prize for The Haloarcula Large Ribosomal Subunit Structure
For this landmark structure, Thomas A. Steitz of Yale University shared the the 2009 Nobel Prize in Chemistry[1] along with two other structural biologists working on the ribosome. It is important to note that the Steitz lab worked with the Moore lab on this phenomenal accomplishment although the Nobel committee limits the award itself to up to three laureates. This structure ranks among the known structures with highest impact.
Haloracula Large Ribosomal Subunit Components
| |||||||
·· ·· ·· ·· ·· ·· ·· |
The large subunit of the Haloracula marismortui ribosome sediments at 50S, as do the large subunits of archaea and eubacteria. It is composed of two chains of RNA, a 23S chain (2,922 nucleotides long, 946 kDa) and a 5S chain (122 bases long, 39 kDa). Assembled with the RNA are 27 protein chains (of a total of 31 known), varying in length from 49 (L39E, 6 kDa) to 337 amino acids (L3, 37 kDa).[2]
The Haloarcula Large Ribosomal Subunit Structure in detail
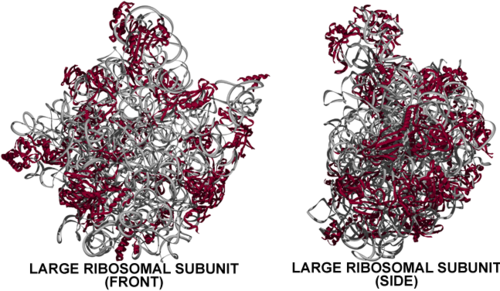
The Haloarcula large ribosomal subunit at first glance:
-
- With no significant portion of the 50S subunit appearing topologically separate or capable of forming a stable structure on its own, the solved structure large subunit is one massive domain.
- On the other hand, the large subunit's partner in translation, the small subunit (30S), clearly has three domains.
- It is important to note that two stalks (the L1 stalk and L7/L12 stalk) seen in lower resolution structures on each side of the large ribosomal subunit at lower resolution [3] are not visible in the higher resolution structure viewed here. Thus, in actuality the Haloarcula large subunit has a less monolithic appearance with protuberances on either side of the central protuberance, yet clearly not possessing the distinct domains formed by the distinct rRNA domains visible in the secondary structure (see below).
- is a ribonucleoprotein macromolecule.
- This macromolecule is mostly RNA sprinkled with several proteins.
- 27 of the 31 known large subunit proteins are visible in the crystal structure. L1, which would be at the 'L1 stalk', is one of the proteins not seen.
- two RNA chains:
- a small 5S rRNA (122 nucleotides)which forms part of the central protuberance seen in the large subunit.
- a large 23S rRNA (3045 nucleotides) - 2833 of the 3045 nucleotides of the 23S rRNA are seen in the structure.
The proteins:
| |||||||
·· ·· ·· ·· ·· ·· ·· |
-
- Proteins are generally globular. However, while about half the proteins seen in the crystal structure of the large ribosomal subunit are globular (shown as orange), interestingly, the other half are extended or have large extended regions emanating from globular domains (shown as cyan).
- These extended proteins and regions are reminiscent of the intrinsically unfolded proteins that play roles in many other processes.
- Zooming in to see some examples in more detail:
- L2, L15, and L39e illustrate proteins with extended regions.
- For contrast, the globular L7ae is shown. The extended regions are highlighted in cyan.
- L39e is extended over its entire length.
-
- View of L2, L15, L39e and L7ae again but with the RNA backbone shown. The extended regions are again highlighted in cyan.
- Using the mouse to spin around the structure clearly shows that the extended proteins penetrate into the interior to fill gaps between RNA secondary structure elements.
- The globular domains are the portions of the proteins on the subunit's exterior, nestling in the gaps and crevices of the folded RNA. You may need to use the mouse to move the structure around to convince yourself.
- This view is also a good point to note the fact that the ribosomal proteins do not encase the nucleic acid as with spherical viruses, nor do the proteins become surrounded by the nucleic acid as in the nucleosome.
Polypeptide Exit Tunnel
- Polypeptide Exit Tunnel
The Peptidyl Transferase Is A Ribozyme
The formation of peptide bonds occurs in the large subunit where the acceptor-stems of the tRNAs are docked.
The initial determination of the atomic resolution structures of the subunits surprisingly revealed that RNA, but not protein, contributes directly to forming the site of both decoding and catalysis of peptide bond synthesis, with the ribosomal proteins only acting in an ancillary role. During the elongation stage of translation, new peptides are added to the carboxy-terminus of the growing nascent chain that is linked to the acceptor-end of the tRNA in the peptidyl or P site. As the nascent chain grows, it advances into a tunnel that passes through the large subunit, called the polypeptide exit tunnel. Several factors can interact at the site of extrusion of the nascent polypeptide chain to ensure proper folding or transport across a membrane. Additionally, during protein synthesis, many additional factors such as elongation factors (EF-Tu and EF-G) interact with the ribosome to elicit decoding and peptide bond synthesis accurately and efficiently. Structures of several of these factors in complex with the ribosome, as well as intermediate states in the process, are being observed now, building upon the first atomic structures.
Structures
Steitz and Moore lab original atomic-resolution structures[4][5]: Haloarcula marismortui large ribosomal subunit - 1ffk and later refined to give 1jj2[6], and then refined to give 1s72[7]. Related: 1ffz, 1fg0. Assembled with the ribosomal RNAs (2,922 and 122 nucleotides long) in the structure are 27 protein chains (of a total of 31 known), varying in length from 49 (L39E, 6 kDa) to 337 amino acids (L3, 37 kDa).[8]
See Also
- Ribosome
- Nobel Prizes for 3D Molecular Structure
- Extremophiles
- Highest_impact_structures of all time
References
- ↑ Nobel Prizes for 3D Molecular Structure
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. PMID:10937989
- ↑ Ban N, Freeborn B, Nissen P, Penczek P, Grassucci RA, Sweet R, Frank J, Moore PB, Steitz TA. A 9 A resolution X-ray crystallographic map of the large ribosomal subunit. Cell. 1998 Jun 26;93(7):1105-15. PMID:9657144
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. PMID:10937989
- ↑ Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000 Aug 11;289(5481):920-30. PMID:10937990
- ↑ Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001 Aug 1;20(15):4214-21. PMID:11483524 doi:http://dx.doi.org/10.1093/emboj/20.15.4214
- ↑ Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004 Jun 25;340(1):141-77. PMID:15184028 doi:10.1016/j.jmb.2004.03.076
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. PMID:10937989
Additional Literature and Resources
- Moore PB. The ribosome returned. J Biol. 2009;8(1):8. Epub 2009 Jan 26. PMID:19222865 doi:10.1186/jbiol103
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009 Oct 29;461(7268):1234-42. Epub 2009 Oct 18. PMID:19838167 doi:10.1038/nature08403
- Ramakrishnan V, Moore PB. Atomic structures at last: the ribosome in 2000. Curr Opin Struct Biol. 2001 Apr;11(2):144-54. PMID:11297922
- RCSB Protein Data Bank coverage of the 2009 Nobel Prizes in Chemistry
- Ribosome: October 2000 Molecule of the Month as part of the series of tutorials that are at the RCSB Protein Data Bank and written by David Goodsell