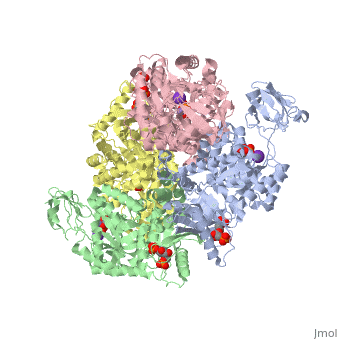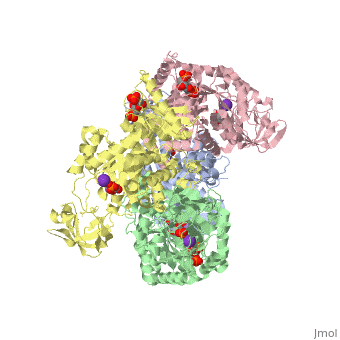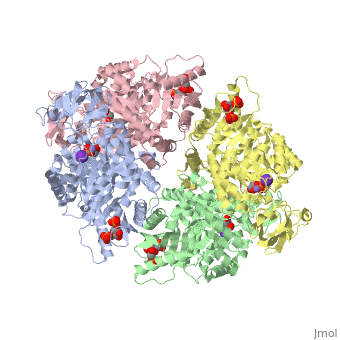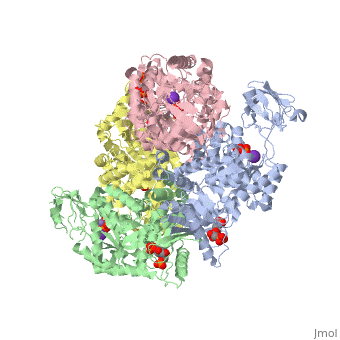
Pyruvate Kinase
From Proteopedia
Contents |
Pyruvate Kinase
Pyruvate kinase is an enzyme that is involved in glycolysis. Pyruvate kinase’s function is to catalyze the last step of glycolysis; thereby, generating the second ATP of glycolysis and pyruvate. It is able to catalyze this step by transferring the phosphate group from phosphoenolpyruvate (PEP) to ADP [1].
| |||||||||
| 2vgb, resolution 2.73Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , | ||||||||
| Activity: | Pyruvate kinase, with EC number 2.7.1.40 | ||||||||
| Related: | 1liy, 1liu, 1liw, 1lix | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Mechanism
[2] catalyzes the final reaction of glycolysis. It couples the free energy of PEP cleavage to the generation of ATP during the synthesis of the final product, pyruvate. This reaction necessitates both K+ and Mg2+ cations, which has two steps. The first step is the nucleophilic attack of the PEP phosphorous atom by β-phosphoryl oxygen of ADP; this step displaces enolpyruvate while forming ATP. In the second step, enolpyruvate tautomerizes to pyruvate [3]. The formation of a high-energy intermediate by enolase in the 9th reaction of glycolysis allows for the synthesis of ATP in this reaction. Though the hydrolysis of 2PG is insufficient in driving the synthesis of ATP, the dehydration of 2PG allows for such a reaction to occur by forming a high-energy intermediate. The high potential of PEP reflects the large release of energy that occurs with the conversion of enolpyruvate to its keto tautomer, pyruvate [4].
Structure
Class: All Beta Proteins
Fold: PK beta-barrel domain-like
Superfamily: PK beta-barrel domain-like
Family: Pyruvate kinase beta-barrel domain
Protein: Pyruvate Kinase (PK)
Species: Human (Homo sapiens)
As can be seen, pyruvate kinase's comprises of in humans. Thus, this enzyme is tetrameric with on each domain for the K+ and Mg2+ ligands to bind to. There are four types of tissue-specific isozymes: L (liver), R (red blood cells), M1 (muscle, heart, and brain), and M2 (early fetal tissue)[5].
Kinetics and Regulation
In the glycolytic cycle, there are three compounds that have a large negative ∆G which includes the reaction pyruvate kinase catalyzes. Due to these three steps regulating the overall activity of the cycle, they are generally irreversible in vivo. Through numerous researches, the activity of pyruvate kinase has been found to be regulated by these effectors:
a. Phosphoenolpyruvate, the substrate, can impact enzymatic activity by enhancing the reaction by allowing the process to operate faster with more substrate present.
b. ATP and pyruvate has been found to be a negative allosteric inhibitor.
c. Alanine has also been found to be a negative allosteric modulator.
As indicated earlier, phosphoenolpyruvate can enhance the activity of the reaction by adding into the enzyme because it is the rate limiting step. The enzyme follows hyperbolic kinetics. Experiments found that no incorporation was found in the reaction, indicating a random, rapid dissociation of the products. This, then, assumes that the products inhibit the enzyme’s reaction by simply reversing the reaction. Both pyruvate and ATP have been shown to be non-competitive inhibitors of pyruvate kinase.
References
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ Robergs, Robert. "Exercise-Induced Metabolic Acidosis: Where do the Protons come from?". 2009. 2/27 2010. <http://www.sportsci.org/jour/0102/rar.htm>.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: John Wiley & Sons, Inc., 2008, 501-503.
- ↑ authors, The scop. "Structural Classification of Proteins". 2009. 2/26 2010. <http://scop.berkeley.edu/data/scop.b.c.jh.b.b.d.html>.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Keegan Gelvoria, David Canner, Ann Taylor, Andrew Alexander