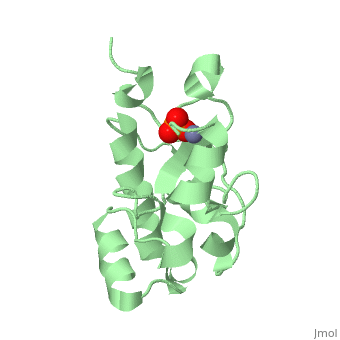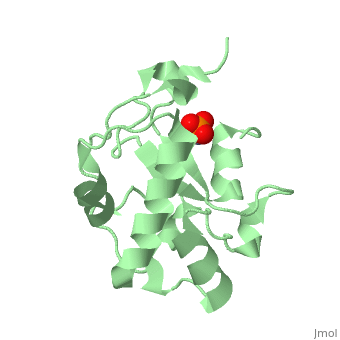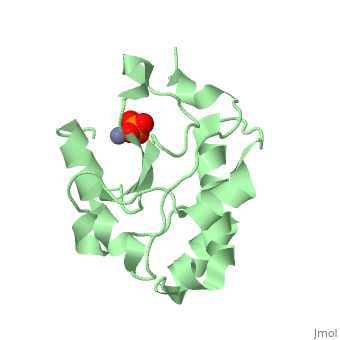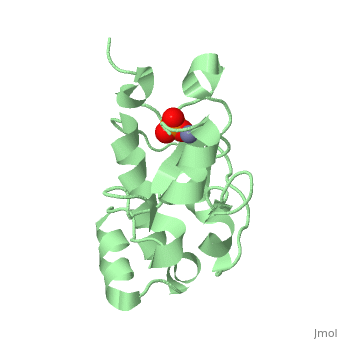
H-N-H motif
From Proteopedia
| |||||||||
| 1fsj, resolution 1.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Deoxyribonuclease I, with EC number 3.1.21.1 | ||||||||
| Related: | 1bxi, 1emv, 1fr2 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The HNH motif is present in many enzymes, including many types of Colicin, such as Colicin E9. It is also at the catalytic centre of homing endonucleases. It adopts ββα-Me structure, where Me indicates the metal ion it binds to.
Proteins containing the HNH motif fall into the HNH superfamily. These include homing endonucleases, colicins, restriction endonucleases, transposases and DNA packaging factors[1]. The core of the HNH domain consists of two antiparallel β-strands connected with a loop of varying length, and flanked by an α-helix, with a metal binding site between the two. In colicins this motif exists independently as an independently folded catalytic domain[2].
The active sites of these nucleases are strongly conserved, and the ability to tolerate a mutation in the conserved catalytic site appears high, and measurable activity is still observed. The activity in the catalytic site is dependent on the presence of a neighbouring imidazole ring, and in the mutations this takes over as a (less efficient) general base for the reaction - usually carried out by the histadine general base. Restrictions on the alternative mutated pathways are probably due to steric contraints placed on the HNH motif, which is involved in protein folding, DNA binding and catalysis simultaneously[3].
The 3 conserved sites (HNH) are strongly structurally and mechanistically conserved across the diverse range of enzymes. The first histadine acts as the general base of the DNA cleavage reaction, and is present at the end of the first β-strand. The asparagine residue stabilises the position of the two β-strands relative to each other. This residue can be replaced by alternative residues that also act to structurally stabilise the position. The final residue is a metal binding histadine, which can be replaced sometimes with a second asparagine instead, creating a HNN motif that acts in the same way as HNH.
Catalytic mechanisms
All studied mechanisms of action have resulted in the same proposed mechanism. The pKa of the histadine general base is elevated due to a nearby hydrogen bond. The imidazole ring, also close by, deprotonates a water molecule and positions it for an in-line nucleophilic attack on the phosphate in the backbone of DNA. A single metal ion bound in the active site stabilises this transition state, but does not participate in the attack[4].
References
- ↑ Cymerman IA, Obarska A, Skowronek KJ, Lubys A, Bujnicki JM. Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease. Proteins. 2006 Dec 1;65(4):867-76. PMID:17029241 doi:10.1002/prot.21156
- ↑ Eastberg JH, Eklund J, Monnat R Jr, Stoddard BL. Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism. Biochemistry. 2007 Jun 19;46(24):7215-25. Epub 2007 May 22. PMID:17516660 doi:10.1021/bi700418d
- ↑ Eastberg JH, Eklund J, Monnat R Jr, Stoddard BL. Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism. Biochemistry. 2007 Jun 19;46(24):7215-25. Epub 2007 May 22. PMID:17516660 doi:10.1021/bi700418d
- ↑ Galburt EA, Chevalier B, Tang W, Jurica MS, Flick KE, Monnat RJ Jr, Stoddard BL. A novel endonuclease mechanism directly visualized for I-PpoI. Nat Struct Biol. 1999 Dec;6(12):1096-9. PMID:10581547 doi:10.1038/70027