Ann Taylor sandbox 117
From Proteopedia
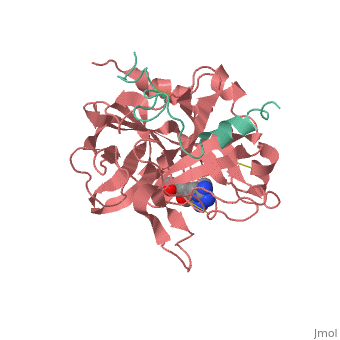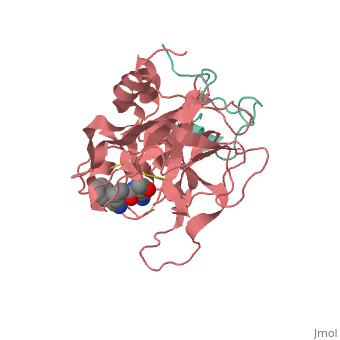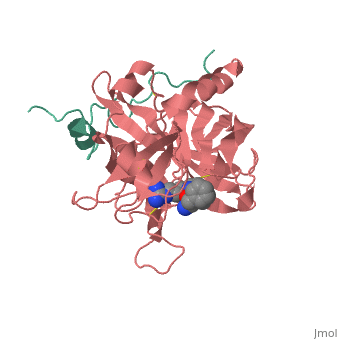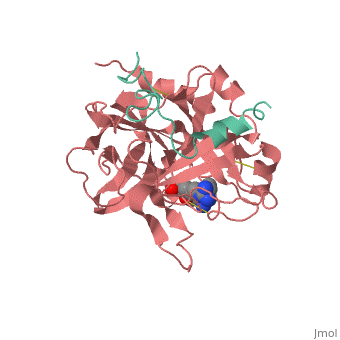
Succinyl-AAPR-trypsin acyl-enzyme
Structures of trypsin acyl-enzymes are used to reconstruct events in the catalytic cycle of serine protease. The structural comparisons provide insight into active site adjustments involved in catalysis. The motions of the catalytic serine and histidine residues coordinated with translation of the substrate reaction center are seen to favor the forward reaction. The structures also clarify how the hydrolytic water attacks in the deacylation reaction. The PDB code is . Movements of the enzyme catalytic residues are subtle but significant: ,
|
Serine proteases, or proteinases, so called due to the presence of a serine residue in the active site, are a class of enzymes that catalyse the hydrolysis of peptide bonds in proteins.
|
Trypsin-BPTI complex
The trypsin backbone is shown in pink and the trypsin inhibitor, BPTI, in yellow (PDB code 2ptc). The residues [Ser195-His57-Asp102-Ser214] are shown in green, the disulfide bond between residues 14-38 is shown in yellow and the Lys 15 sidechain at the specificity site in pink.
Content Donators
- Content for this page has been included and adapted with permission from Jane S. and David C. Richardson's http://kinemage.biochem.duke.edu/
Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load
and display another structure.
| |||||||||
| 2age, resolution 1.15Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Non-Standard Residues: | |||||||||
| Activity: | Trypsin, with EC number 3.4.21.4 | ||||||||
| Related: | 2agg, 2agi, 2ah4
| ||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||