Sandbox Reserved 164
From Proteopedia
| This Sandbox is Reserved from March 9, 2011, through May 30, 2011 for use by the course Biochemistry at Reinhardt University, Waleska, USA, taught by Irma Santoro. This reservation includes Sandbox Reserved 162 through Sandbox Reserved 168. |
To get started:
More help: Help:Editing |
|
Contents |
Superoxide Dismutase 1 (SOD 1)
SOD1 is one of three oxidoreductase enzymes that is responsible for binding copper and zinc ions to highly reactive oxygen free radicals and transforming them into oxygen and hydrogen peroxide [1]. This occurs in a quick two step mechanism
M(n+1)+-SOD + O2− → Mn+-SOD + O2
Mn+-SOD + O2− + 2H+ → M(n+1)+-SOD + H2O2[2].
This protein is coded for by the SOD 1 gene located on chromosome 21 at position 21q22.1 from base pairs 33,031,934 to 33,041,243 [3].
Structure
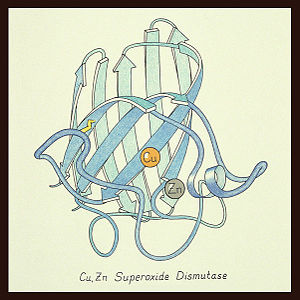
The SOD 1 protein is a homodimer with an amino acid sequence length of 154. SOD 1 has an 8-stranded "Greek key" beta-barrel shape with the active site located between the barrels. The copper and zinc ligands are made up of six histidine side-chains and one aspartate side-chain with the metal ions connected with by a single histidine chain.[2]. This first SOD structure was determined by Irwin Fridovich and Joe McCord in 1973 [1][4] with the SOD 1 "Greek key" structure visualized by Dr. Jane Richardson (see below).[2]
ALS
References
- ↑ 1.0 1.1 McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969 Nov 25;244(22):6049-55. PMID:5389100
- ↑ 2.0 2.1 2.2 Tainer JA, Getzoff ED, Richardson JS, Richardson DC. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983 Nov 17-23;306(5940):284-7. PMID:6316150
- ↑ SOD 1. Genetics Home Reference. U.S. National Library of Medicine; 2010
- ↑ McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968-1988). Free Radic Biol Med. 1988;5(5-6):363-9. PMID:2855736