User:Bianca Varney/Replication Terminator Protein
From Proteopedia
|
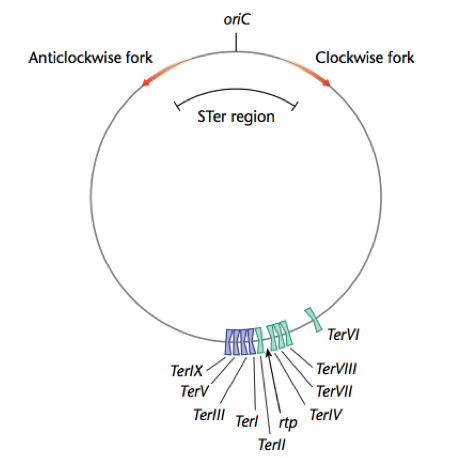
Replication Terminator Protein (RTP) arests replication in gram positive bacteria. RTP binds bacterial DNA at Termination (Ter) sites that lie opposite the origin of replication (OriC). When the replication forks meet with RTP, which bound to Ter sites, replication is arrested, and DNA polymerase falls off the bacterial chromosome. However, RTP-Ter interactions are orientation specific, and will only arrest the replication forks traveling in on one direction; either clockwise or anticlockwise once each fork has copied more than half the bacterial chromosome. This permits the entire bacterial chromosome to be copied.
Contents |
Mechanism
Bacterial replication termination has been well studied in E. coli and B. subtilis. The proteins involved in this termination process differ structurally in these two bacterium, although each contains similar contrahelicase activity and performs similar functions in arresting replication. The replication termini sequences are located in two clusters approximately 180 degrees from the origin of replication, and are orientated such that each cluster has opposite polarity and are therefore inverted repeats. This means that counterclockwise replication forks can move beyond the first cluster of ter sites, the left terminus, but are arrested at the second cluster, the right terminus, containing correct polarity (FIG). Similarly, the clockwise fork will proceed through the left cluster but will be arrested at the right. The bipartite Ter nucleotide sequence is overlapping and each inverted repeat contains a core (IRIB) and an axillary (IRIA) sites. RTP binds to these sequences, resulting in the impediment the replication fork helicase.
Biological Role
Structure
The replication termination protein is a homodimer composed of two 14.5 kDa subunits. Each RTP monomer consists of four α helical and three β strands, an unstructured N-terminal containing six amino acids, and an extended loop. The monomer comprises of a 4-turn α-helix (α1) connected to a β-strand (β1), from the N to C terminus. The β1 strand connects to α3, a 3-turn α-helix, via α2, a 3-turn helix. The α3 helix is attached to a β-pleated sheet. Antiparallel in structure it contains the β2 and β3 strands proximal to the β1 strand. The C-terminal contains the last α helix, the 8-turn α4 helix. The connections between the secondary structures are short, with the exception of the elements connecting α2-->α3 and β2-->β3. The latter is an important structure in dimer-dimer interactions, as it is predicted to contact the structure of an adjacent dimer. This was backed by evidence from site-directed mutagenesis as mutations in this region, specifically residue 34 and 82, results in no dimer-dimer interaction. The loop and strands create a wing, of the winged helix. The winged helix is involved in regions of DNA binding, and analysis has revealed that the RTP protein has structural homology to the H5 histone protein, due to this similar region. Although it is important to note that no significant similarity between sequences was determined between RTP and H5 histone protein. The RTP protein contains three major structural domains for its specific functionality, including DNA-binding, DnaB interaction and dimer-dimer interaction domains. Biochemical and mutational studies have identified particular residues that are vital for the functionality of the RT protein. Manna et al have identified that mutation within a hydrophobic region at residues Glu-30 and Tyr-33 causes the loss of contrahelicase ability. These mutations do not affect dimer-dimer interactions or DNA binding activity and indicate that simple DNA binding is not able to block the replication fork. This provided evidence that RTP and the replication fork machinery interact specifically.
The RTP is organized into a dimer by the association of their long α helices within the C-terminus. The ‘winged helix’ is believed to be involved as the major DNA-binding domain however two of the α helices, at the centre of the protein, and two β strands, in the outer regions, have been suggested to fit adjacently into the major and minor grooves respectively. The unstructured N terminal region is may also have a role in DNA-binding. This binding interaction is different from the Tus-Ter interactions in E. coli.