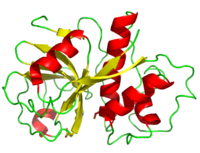
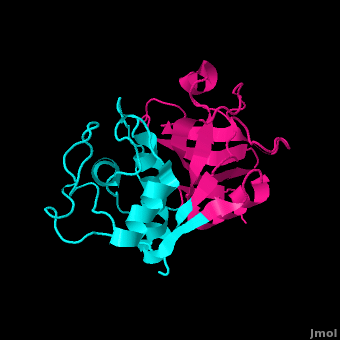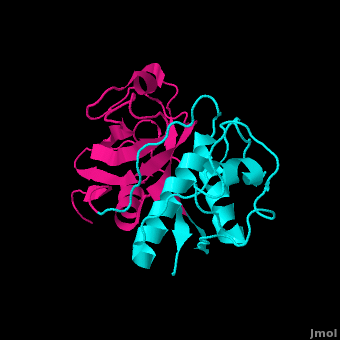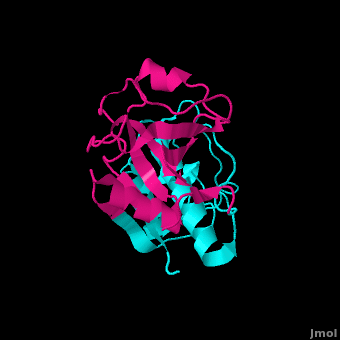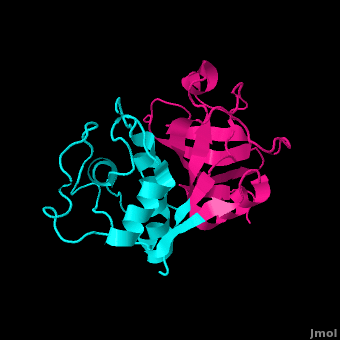
Structure
Contacts between Papain Subunits.
[14].
Papain's two subunits are held together with "arm" linkage where one end of the protein chain holds the opposite domain. In papain's case the "arm crossing" primarily occurs on or near the surface. [14]
As with all proteins, folding to form secondary and tertiary structures is largely determined by the interactions of the to exclude water (hydrophobic residues shown in purple). A majority of these hydrophobic residues form within each of the lobes ensuring their stability (surface residues are transparent, buried hydrophobic residues are opaque and colored). The remaining residues are , some carrying a (acidic) at physiological pH, others a (basic), the rest of the polar residues are neutral. The protein's tertiary structure consists of two domains divided by a cleft in which the active site resides.[19]
The 212 residues of Papain can be split nearly in half to produce though the enzyme consists only of one polypeptide chain.[20] The is located in the cleft between the two domains. The two domains interact with one another via hydrophobic interactions, hydrogen bonds, and electrostatic interactions in this cleft. For example, from the L Domain hydrophobically interacts with the carbon atoms on residues Lys174, Ala162 and Pro129 of the R Domain. hydrogen bonds multiple times with the oxygen atoms of Ser176 and also with the oxygen atom on Tyr88. Electrostatic interactions are seen between where the carboxyl group of Glu35 forms an ionic bond with the ammonia group of the Lys174 residue. The sum total of interactions within the cleft between the two domains ensure that the lobes do not move with respect to one another. [21]
The secondary structure of papain consists of 7 , 17 , all of which are antiparallel, and a large amount (about 50% of total residues) of . The , which goes from blue (amino terminus) to red (carboxyl terminus) is useful for tracing the order of these structures through the chain. These secondary structures form as a result of favorable hydrogen bonding interactions within the polypeptide backbone. Meanwhile, secondary structures are kept in place by hydrophobic interactions and hydrogen bonds between sidechains of adjacent structures. For example, the (residues 25-42) is maintained as a result of between backbone carbonyl atoms and the hydrogen on the amide nitrogen four residues away. However, are present between this helix and the rest of the protein, suggesting that this helix is coordinated primarily by hydrophobic interactions. This is reasonable given its central location in the enzyme. As expected, the helix contains many (red residues are hydrophilic). The tertiary structure of papain is also maintained by three , which connect , , and [8]. These disulfide bonds are likely important in conserving the structural integrity of the enzyme as it operates in extracellular environments at high temperatures.
The active site contains a made up of Cysteine-25 and Histidine-159. Aspartate-158 also plays a role in catalysis but it is not considered part of the diad. Papain's active site can accommodate seven amino acids of a substrate. When the peptide is cleaved, the first four resides reside on the amino side of the peptide bond while the other three reside on the carboxyl side. [22] Papain prefers to cleave at: (hydrophobic)-(Arg or Lys)- cleaves here -(not Val). Hydrophobic is Ala, Val, Leu, Ile, Phe, Trp, or Tyr. [23]
Catalytic Mechanism
Like serine proteases, cysteine proteases contain a of residues. In the case of papain, these residues are Cys-25, His-159, and Arg-175. Papain also contains a fourth residue, that has been shown to play an important role in catalysis and is likely involved in the formation of the oxyanion hole. The mechanism begins when a peptide binds to the active site. Papain's Cys-25 is deprotonated by its His-159. The now nucleophilic Cys-25 attacks the carbonyl carbon of the peptide backbone, forming an acyl enzyme intermediate in which the peptide's amino terminal is free. Also in this step, His-159 is returned to its deprotonated form. The intermediate is then deacylated by a water molecule, and it releases the carboxyl terminal of the peptide to produce the product and regenerate the active enzyme. This entire mechanism is shown below:
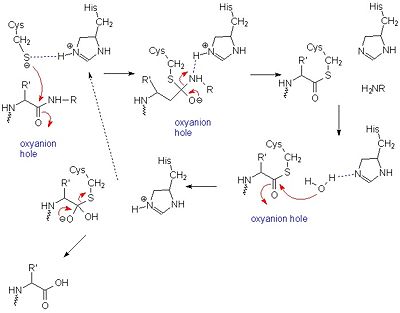
General mechanism of papain catalysis
[24]. Arg-175, which orients His 159, and Gln-19, which contributes to the formation of the oxyanion hole, are not shown.
[25].
Papain is also known to cleave antibodies above and below the disulfide bonds that join the heavy chains and that is found between the light chain and heavy chain. This generates two monovalent Fab segments, that each have a single antibody binding sites, and an intact Fc fragment, as shown below:
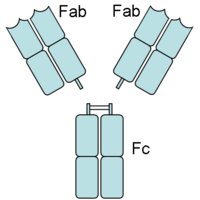
Antibody Cleavage by Papain
[26]]]
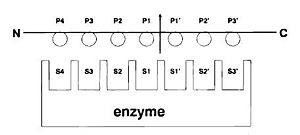
Papain has a broad specificity for protein substrates. The active site consists of seven subsites (S1-S4 and S1’-S3’) that can each accommodate one amino acid residue of a protein substrate (P1-P4 and P1’-P3’).
[27]]]
Catalytic Inhibitors
acts as a competitive inhibitor to cysteine proteases-- it binds tightly but reversibly to the papain active site. Stefin inhibitors are characterized by Mr of about 11,000, no disulfide bonds and no associated carbohydrates.
In Stefin B, the Gly9 residue along with two hairpin loops form a "wedge" complementary to the active site groove of papain. This wedge makes extensive and tight interactions with papain and a total of 128 intermolecular atom-atom interactions occur. Met6-Pro11, Gln53-Asn59, Gln101-His104 and Tyr124-Phe125 on the wedge all have some interaction to the enzyme though not always direct. All residues from the base and both sides of the are involved in the complex with the inhibtor (Trp177, Ser21, Cys63, Cys25, Asp158 and His159).
There are a small number of between stefin B and papain, however there are many more polar interactions mediated by . Thirteen solvent molecules bridge polar residues of the enzyme and inhibitor. Seventeen hydrogen bonds are made with a solvent molecule and stefin. Fourteen of these bridges form a papain contact. The rest of the interactions are largely hydrophobic-- involving apolar . [28]
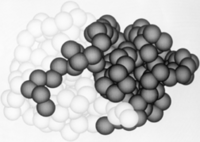
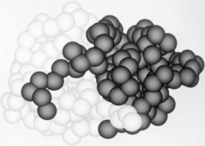
Leupeptin (PDB ID #: 1pop)
is a commonly studied inhibitor of proteases (seen in the Jmol as ball and stick model). It inhibits by binding and interacting with the active site which allows it to block the enzyme's desired protein substrate. There are many that interact with Leupeptin in the active site. The predominant interaction is from hydrophobic interactions between Leupeptin and . In addition to hydrophobic interactions, there are also some hydrogen bonding interactions to hold Leupeptin in the active site of papain. Leupeptin works well at blocking papain from its enzymatic duties. A recent study has shown that Leupeptin actually forms a covalent bond between its and CYS 25. In addition, the residues Gln 19 and CYS 25 form with the Leupeptin molecule
In order to investigate binding of protein substrates to papain, Schröder et. al. crystallized the enzyme with the broad-spectrum competitive protease inhibitor leupeptin, shown in blue in the ribbon diagram. It has the structure Ac-Leu-Leu-Arginal, where Ac is an acetyl group attached to the nitrogen of the first leucine. The inhibitor functions by binding to the enzyme's active site, where the catalytic nucleophile (cysteine in papain) attacks the arginal aldehyde. This forms a tight-binding transition state from which the normal catalytic mechanism cannot proceed, due to this carbonyl having no potential leaving groups bonded to it. Analysis of the resulting structure revealed that the of papain consists primarily of a variety of , including tyrosine, tryptophan, and valine, which coordinate the bound leuptin. Some of the enzyme's residues also make with some of the leupeptin atoms. These hydrogen bonds, shown in yellow, include interactions between both hydrogens on both Gln-19 and the amide nitrogen of the catalytic Cys-25 with the arginal carbanion, forming the catalytically important oxyanion hole. In addition, Gly-66 interacts with the second leucine in leupeptin while Asp-158 interacts with a hydrogen on the arginal. These interaction further stabilize and orient the substrate in the binding pocket[29].
Medicinal Uses
Papain has been used for a plethora of medicinal purposes including treating inflammation, shingles, diarrhea, psoriasis, parasites, and many others.[30] One major use is the treatment of cutaneous ulcers including diabetic ulcers and pressure ulcers.[31] Pressures ulcers plague many bed bound individuals and are a major source of pain and discomfort. Two papain based topical drugs are Accuzyme and Panafil, which can be used to treat wounds like cutaneous ulcers.[32]
A recent New York Times article featured papain and other digestive enzymes.[34] With the number of individuals suffering from irritable bowel syndrome and other gastrointestinal issues, many people are turning toward natural digestive aid supplements like papain. The author even talks about the use of papain along with a pineapple enzyme, bromelain, in cosmetic facial masks. Dr. Adam R. Kolker (a plastic surgeon) is quoted in the article saying that "For skin that is sensitive, enzymes are wonderful." He bases these claims off the idea that proteases like papain help to break peptide bonds holding dead skin cells to the live skin cells.[35]
Papain digests most proteins, often more extensively than pancreatic proteases. It has a very broad specificity and is known to cleave peptide bonds of basic amino acids and leucine and glycine residues, but prefers amino acids with large hydrophobic side chains. This non-specific nature of papain's hydrolase activity has led to its use in many and varied commercial products. It is often used as a meat tenderizer because it can hydrolyze the peptide bonds of collagen, elastin, and actomyosin. It is also used in contact lens solution to remove protein deposits on the lenses and marketed as a digestive supplement. Another place papain has found use is clinical medicine, where it is used to treat pain, swelling, and fluid retention following trauma and surgery. [36] Finally, papain has several common uses in general biomedical research, including a gentle cell isolation agent, production of glycopeptides from purified proteoglycans, and solubilization of integral membrane proteins. It is also notable for its ability to specifically cleave IgG and IgM antibodies above and below the disulfide bonds that join the heavy chains and that is found between the light chain and heavy chain. This generates two monovalent Fab segments, that each have a single antibody binding sites, and an intact Fc fragment, as shown in the image to the right:[11]
Reference
- Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984 Oct 25;179(2):233-56. PMID:6502713
Image from: http://upload.wikimedia.org/wikipedia/commons/5/5c/Cysteinprotease_Reaktionsmechanismus.svg
Image from: http://en.wikipedia.org/wiki/Papain
Page seeded by OCA on Tue Feb 17 04:20:31 2009