Sandbox 35
From Proteopedia
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
Contents |
Papain
Introduction
Did you know?
. Meat tenderizer. Old time home remedy for insect, jellyfish, and stingray stings[1]. Who would have thought that a sulfhydryl protease from the latex of the papaya fruit, Carica papaya and Vasconcellea cundinamarcensis would have such a practical application beyond proteopedia?
This protease belongs to an extended family of aminopeptidases, dipeptidyl peptidases, endopeptidases, and other enzymes having both exo- and endo-peptidase activity. The inactivated zymogen with N-terminal propeptide regions - providing stability in alkaline environments and enabling proper folding - is activated through removal of the propeptide regions. [2] The protein is primarily secreted with its pro-region enabling transport from zymogen to lysosome through membrane association and mediation. [3]
Historicity
Papain made its first appearance in the Calcutta Medical Journal entitled “The Solvent Action of Papaya Juice on Nitrogenous Articles of Food” when G.C Roy was investigating the enzyme in 1873. In the late 19th century, Wurtz and Bouchut dubbed the partially purified enzyme "papain." [4] At the time, it was viewed as proteolytically active constituent in the latex of tropical papaya fruit. [5] As separation and purification techniques improved, pure papain was able to be isolated. In becoming the second enzyme to attain an X-ray crystallized structure and the first cysteine protease to behold an identifiable structure, papain fueled greater advances in enzymatic studies. [6]
Papain. Lights. Camera. Action!
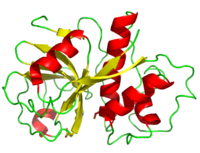
Structure
| |||||||||||
Catalytic Mechanism
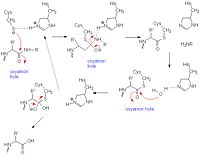
Papain's catalytic mechanism is like serine proteases. Its catalytic triad of residues Cys 25- His159- Asn-175 appear to work with a fourth residue, Gln-19, suspected to be involved in oxyanion hole formation. When a peptide binds to the active site, His-159 deprotonates Cys-25 which in turn attacks the substrate carbonyl carbon. The oxyanion hole then stabilizes the resulting covalent, tetrahedral intermediate. Subsequently, nitrogen in the peptide bond is protonated by His-159 (acting as an acid). This action frees the C-terminal portion of the peptide so that it is released. The entrance of water into the active site then attacks the carbonyl carbon while it is deprotonated by His-159, resulting in another tetrahedral covalent intermediate once again stabilized through the oxyanion hole. At the end, carbonyl reformation and the Cys-25 sulfur action as the leaving group releases the N-terminal portion of the peptide. The enzyme is regenerated for the cycle to begin again. [18]
Other Interaction with Inhibitors and Effectors
Except for valine, papain prefers to cleave at hydrophobic residues alanine, leucine, isoleucine, phenylalanine, tryptophan, or tyrosine [20]. Because of the importance of the oxyanion hole formation and the nucleophilic attack of cysteine, substances like cysteine, sulfide/sulfite, heavy metal chelating agents like EDTA, and N-bromosuccinimide behave as activators of the enzyme while PMSF, Hg2+ and other heavy metals, cystatin, leupeptin, sulfhydryl binding agents, carbonyl reagents, and alkylating agents serve as inhibitors. [21][22]
Fun Trivia
Remember the 2002 SARS (Severe Acute Respiratory Syndrome) epidemic that placed global health, particularly in Southeast Asia, in a precarious state? On-going research is happening to further understand the mechanisms of this coronavirus, so that future steps can be taken for prevention. Its been found that the replication of RNA for this virus is mediated by two viral proteases that have many papain-like characteristics. [23]
Despite a low percentage of sequence identities, inhibition and sequence analyses have increasingly been drawing parallels between L proteinases, that involve the foot-and-mouth disease virus and equine rhinovirus 1, and papain. With a similar overall fold to papain and identifiable regions that resemble the five alpha-helices and seven beta-sheets of papain, L proteinases of foot-and-mouth disease virus and of equine rhinovirus 1 reveal a mode of operation that is very papain like. [24]
References
- ↑ [1] Ameridan International
- ↑ Rawlings ND, Barrett AJ. Families of cysteine peptidases. Methods Enzymol. 1994;244:461-86. PMID:7845226
- ↑ Yamamoto Y, Kurata M, Watabe S, Murakami R, Takahashi SY. Novel cysteine proteinase inhibitors homologous to the proregions of cysteine proteinases. Curr Protein Pept Sci. 2002 Apr;3(2):231-8. PMID:12188906
- ↑ Menard and Storer 1998
- ↑ Wurtz and Bouchut 1879
- ↑ Drenth J, Jansonius JN, Koekoek R, Swen HM, Wolthers BG. Structure of papain. Nature. 1968 Jun 8;218(5145):929-32. PMID:5681232
- ↑ [2]9PAP PDB
- ↑ 8.0 8.1 8.2 8.3 Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984 Oct 25;179(2):233-56. PMID:6502713
- ↑ 9.0 9.1 [3] Jane S. Richardson
- ↑ Schechter and Berger 1967
- ↑ Kimmel and Smith 1954
- ↑ 12.0 12.1 Wang J, Xiang YF, Lim C. The double catalytic triad, Cys25-His159-Asp158 and Cys25-His159-Asn175, in papain catalysis: role of Asp158 and Asn175. Protein Eng. 1994 Jan;7(1):75-82. PMID:8140097
- ↑ Ménard R, Khouri HE, Plouffe C, Dupras R, Ripoll D, Vernet T, Tessier DC, Lalberté F, Thomas DY, Storer AC. A protein engineering study of the role of aspartate 158 in the catalytic mechanism of papain. Biochemistry. 1990 Jul 17;29(28):6706-13. PMID:2397208 doi:10.1021/bi00480a021
- ↑ [4] The Journal of Biological Chemistry
- ↑ [5] WebMD
- ↑ Tsuge H, Nishimura T, Tada Y, Asao T, Turk D, Turk V, Katunuma N. Inhibition mechanism of cathepsin L-specific inhibitors based on the crystal structure of papain-CLIK148 complex. Biochem Biophys Res Commun. 1999 Dec 20;266(2):411-6. PMID:10600517 doi:10.1006/bbrc.1999.1830
- ↑ Skern T, Fita I, Guarne A. A structural model of picornavirus leader proteinases based on papain and bleomycin hydrolase. J Gen Virol. 1998 Feb;79 ( Pt 2):301-7. PMID:9472614
- ↑ 18.0 18.1 [6] University of Maine
- ↑ [7] Worthington Biochemical Corporation
- ↑ [8] Sigma Aldrich Papain
- ↑ Nicklin MJ, Barrett AJ. Inhibition of cysteine proteinases and dipeptidyl peptidase I by egg-white cystatin. Biochem J. 1984 Oct 1;223(1):245-53. PMID:6388564 doi:10.1042/bj2230245
- ↑ [9] Biozym
- ↑ Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005 Dec;79(24):15189-98. PMID:16306590 doi:10.1128/JVI.79.24.15189-15198.2005
- ↑ Skern T, Fita I, Guarne A. A structural model of picornavirus leader proteinases based on papain and bleomycin hydrolase. J Gen Virol. 1998 Feb;79 ( Pt 2):301-7. PMID:9472614