SandboxPKA
From Proteopedia
Contents |
Introduction
|
The c-Abl protein 1 (ABL1), also known as Abelson kinase, is a non-receptor tyrosine kinase that plays a role in many key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, DNA damage response and apoptosis. [1] [2] Activity of c-Abl protein is negatively regulated by its SH3 domain, and deletion of the SH3 domain turns ABL1 into an oncogene. In more than 90% cases, chronic myelogeneous leukemia (CML) is caused by chromosomal abnormality resulting in the formation the Philadelphia chromosome. This chromosome is formed by fusion between Abelson (Abl) tyrosine kinase gene at chromosome 9 and break point cluster (Bcr) gene at chromosome 22, resulting in the chimeric oncogene Bcr-Abl and a constitutively active Bcr-Abl tyrosine kinase. Small molecule inhibitors of Bcr-Abl that bind to the kinase domain can be used to treat CML [3]
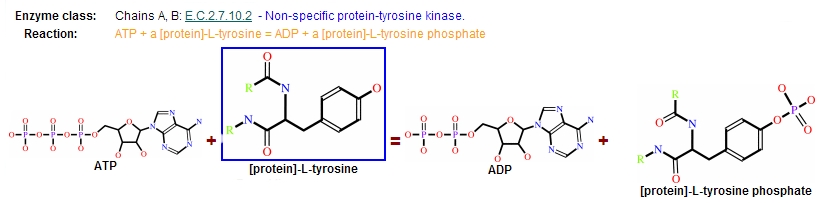
Reaction
Protein kinases are a group of enzymes that possess a catalytic subunit that transfers the gamma (terminal) phosphate from nucleotide triphosphates (often ATP) to one or more amino acid residues in a protein substrate side chain, resulting in a conformational change affecting side protein function.
The enzymes are classified into two broad groups, characterised with respect to substrate specificity:
- Serine/threonine kinases
- Tyrosine specific kinases: c-Abl is included in this group [4]
Structure
All of the protein kinases have a similar bilobal fold, and their key structural features have been well studied
| |||||||||||
Catalytic domain
It is responsible of both, ATP binding as well as protein binding.
| |||||||||||
Resistance
In the majority of cases, resistance is caused by reactivation of Bcr–Abl kinase activity. Mutations in the BCR–ABL gene, predominantly in the kinase domain, are the most common mechanism implicated in secondary drug resistance. In some treated patients, BCR–ABL gene amplification and increased Bcr–Abl protein expression have been observed as a compensatory mechanism for the imatinib antitumour effect. The switch to alternative kinase signalling pathways, which can compensate for the loss of Bcr–Abl activity, has been proposed as another resistance strategy of leukaemic cells. Imatinib might also have different effects on chronic myelogenous leukaemia (CML) tumour cells depending on their differentiation state, and it has been proposed that quiescent CML stem cells might be intrinsically resistant to the drug. Other mechanisms of resistance to imatinib might be related to pharmacokinetic factors of drug delivery. Imatinib can be actively transported out of tumour cells through efflux pump proteins to keep intracellular drug concentrations below inhibitory levels. Extracellular sequestration of imatinib by 1 acid glycoprotein in the plasma has been proposed as a potential mechanism, which would result in reduced availability of the free drug to CML cells [5] .
References
- ↑ http://www.ncbi.nlm.nih.gov/pubmed/9037071
- ↑ http://www.ncbi.nlm.nih.gov/pubmed/11114745
- ↑ Crystal Structures of the Kinase Domain of c-Abl in Complex with the Small Molecule Inhibitors PD173955 and STI571
- ↑ Leukemia research 34 (10): 1255–1268. doi:10.1016/j.leukres.2010.04.016. PMID 2053738
- ↑ http://www.nature.com/nrd/journal/v3/n12/full/nrd1579.html