Sandbox 126
From Proteopedia
Jump to: navigation, search
proteopedia linkproteopedia link
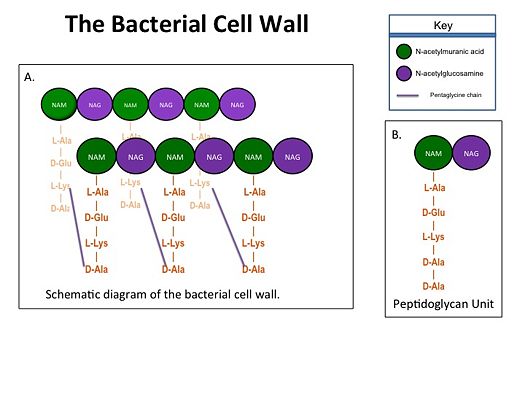
BackgroundTranspeptidases (TP), also known as penicillin-binding proteins (PBP), catalyze the cross-linking of peptidoglycan polymers during bacterial cell wall synthesis. The natural transpeptidase substrate is the D-Ala-D-Ala peptidoglycan side chain terminus. Beta-lactam (β-lactam) antibiotics, which include penicillins, cephalosporins and carbapenems, bind and irreversibly inhibit transpeptidases by mimicking the D-Ala-D-Ala substrate, resulting in the inhibition of cell wall synthesis and ultimately bacterial cell growth. Overuse and misuse of β-lactams has led to the generation of methicillin- resistant Staphylococcus aureus (MRSA) isolates that have acquired an alternative transpeptidase, PBP2a, which is neither bound nor inhibited by β- lactams. MRSA isolates are resistant to all β-lactams, and are often only susceptible to “last resort antibiotics”, such as vancomycin. Recently, two cephalosporins - ceftobiprole and ceftaroline - that bind and inhibit PBP2a have been developed.
How does PBP2a workPBP2a is composed of two domains: a The NBP domain of PBP2a is anchored in the cell membrane, while the TP domain “sits” in the periplasm with its active site facing the inner surface of the cell wall. The active site contains a serine residue at position 403 (Ser403) which catalyzes the cross-linking of the peptidoglycan rows with pentaglycine cross-links. | |||||||||||
Retrieved from "http://52.214.119.220/wiki/index.php/Sandbox_126"