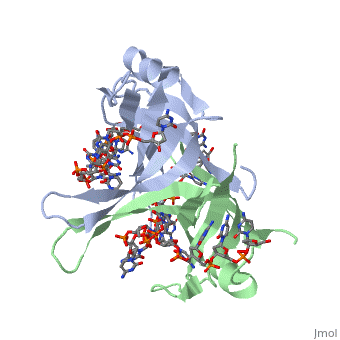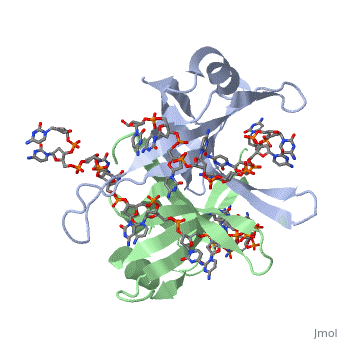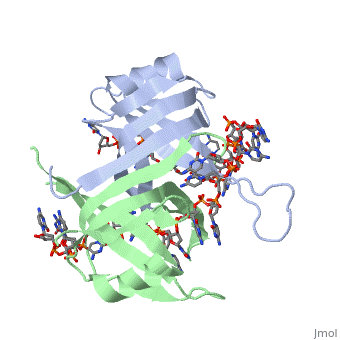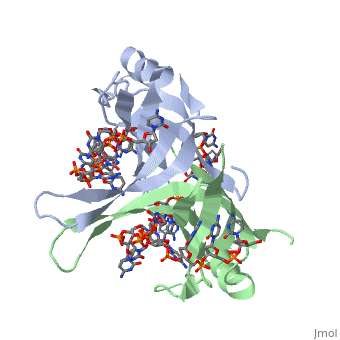
SSB consists is a homotetramer that has a DNA binding domain which binds to a single strand of DNA. The tetramers consist of α-helices, β-sheets, and random coils. Each subunit contains an and several . The secondary structure also includes a NH2 terminus, which consists of multiple positively charged amino acids. The DNA-binding domain lies within 115 amino acid residues from this terminus [2]. The COOH terminus includes many acidic amino acids.
Binding Interactions in the Active Site
ssDNA can interact with binding proteins through hydrogen bonds, stacking, or electronegative interactions. Most interactions between SSB and ssDNA happen through the OB fold. OB stands for oligosaccharide/oligonucleotide binding site. This fold consists of a 5-stranded β barrel that ends in an α-helix.
is an important DNA binding site. It has been shown to be the site for cross-linking.
Tryptophan and Lysine residues are important in binding as well. Treatments resulting in
modification of arginine, cysteine, or tyrosine residues had no effect on binding of SSB to
DNA, whereas modification of either lysine residues (with acetic anhydride) or tryptophan
residues (with N-bromosuccinimide) led to complete loss of binding activity [3].
SSB-Protein Interactions
SSB can form complexes with many other proteins. This trait can keep enzymes needed for damage repair, transcription, etc. near the ssDNA and it is thought that SSB can even help to stimulate these enzymes to carry out their jobs. When DNA binds SSB, most of the molecule loses flexibility. But the COOH terminal domain remain flexible, even after DNA binding. It is believed that the COOH terminus has something to do with protein binding [4].
SSB will interact with the protein RecA to enable recombination, because RecA will recognize SSB and replace it on the strand.
In DNA repair, SSB will bind to the damaged strand to protect it. And eventually it will attract repair enzymes which will replace SSB and begin repair mechanisms.
SSB has also been thought to bind with exonuclease I, DNA polymerase II,
and a protein n, which is used to help synthesize RNA primers for the lagging strand. SSB can also help regulate transcription by competing with other proteins for binding spaces on DNA. SSB has a higher affinity for DNA than most other proteins, and those proteins are not able to remove SSB from DNA and bind themselves. This type of mechanism can not only regulate transcription, but it can provide protection for the DNA [5].