Introduction
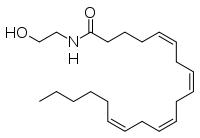
Fatty acid amide hydrolase (FAAH) degrades fatty acid amides to terminate their signaling activity.(IMT5) A serine hydrolase from the Amidase signature superfamily of enzymes (other amidases), FAAH degrades endocannabinoid signaling lipids, molecules associated with pain relief.(2VYA) Because endocannabinoids are lipid molecules, they cannot be compartmentalized in vesicles (the degradation method for other neurotransmitters) and must instead be degraded in the bilayer of the cell membrane. FAAH is an integral membrane protein that degrades FAAs as they enter the membrane bilayer. (IMT5) Current research about FAAH aims to find inhibitors for the enzyme, which would prolong the pain alleviation provided by endocannabinoid molecules. (2VYA)
Hydrolase Information
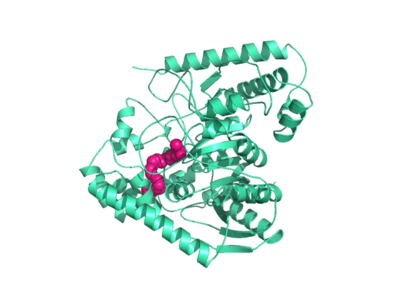
Crystal structures of FAAH show that the enzyme is a homodimer in solution, with each subunit having a mass of 63 kD. The protein's of 11 strands is surrounded by 24 alpha helices. The enzyme is embedded in the cell to catch the lipid signaling molecules that can diffuse through membranes. The FAAH structure shows an entry channel leading from the lipid bilayer to the enzyme's active site, providing a path for endocannabinoids to enter the hydrolase. In addition, FAAH possesses a channel leading from the active site to the cell's cytoplasm, allowing the release of polar compounds released from lipid cleavage and the entry of water molecules necessary for the FAAH mechanism to proceed. (IMT5)
This hydrolase has a membrane binding cap, a consisting of alpha helices 18 and 19. These helices present hydrophobic amino acid residues that likely help FAAH interact with the hydrophobic region of the lipid bilayer. (IMT5)
with
Catalytic Triad
Mutagenesis and inhibitor studies have shown that FAAH has a , consisting of S241, S217, and K142. Ser-Ser-Lys catalytic triads are not often seen in hydrolases, making FAAH an enzyme of interest for additional research. S241 acts as the catalytic nucleophile for the cleavage of amide bonds. (IMT5)
FAAH requires two water molecules in its active site to properly cleave amide bonds. One water molecule (W1) deacylates the substrate, and the other (W2) helps coordinate W1 through the catalytic K142. (3LJ6)