You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
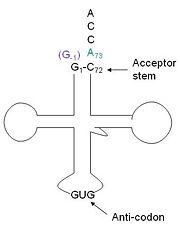
tRNA His, highlighting the incoming G-1 (purple) opposite A73 (green)
a. tRNA His (link to wikipedia) has a guanine monophosphate (GMP) residue at the 5’ end in all domains of life, besides α-proteopbacteria. This GMP is referred to as G-1. In prokaryotes (link to wikipedia) G-1 is encoded in the genome. RNase P (link to wikipedia) cleaves pre-tRNAHis to generate the mature tRNA, leaving an extra basepair on the acceptor stem, G-1:C73. In eukaryotes the G-1 residue is not encoded and needs to be added post-transcription. The enzyme that catalyzes this reaction is the polymerase, tRNAHis guanylyltransferase (Thg1). Howerver, the addition of the GMP residue is nontemplated, inserting GMP across from A73 in the acceptor stem creating a mismatch. Unlike most polymerases, Thg1 adds nucleotides in the 3’ –to- 5’ direction, while forming a normal 3’ –to- 5’ phosphodiester bond. Therefore, the 3’-OH of the incoming nucleotide attacks the 5’ end of the polynucleotide chain. This is a two step mechanism where the polynucleotide chain is first adenylated and then guanylated.
Classification
Structure
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.

