Sigma factor
From Proteopedia
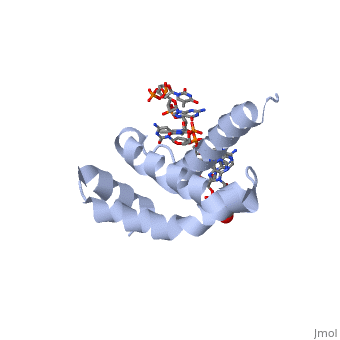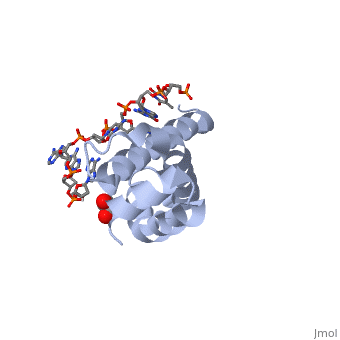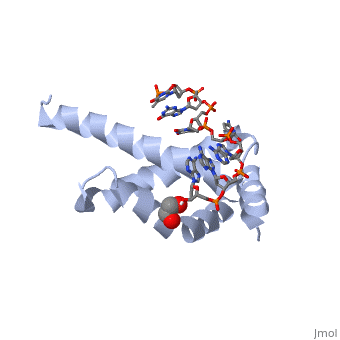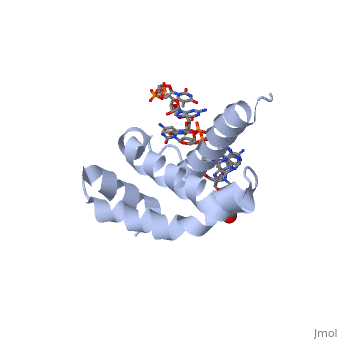
OverviewSigma (σ) factor is the peoptide subunit needed for the initiation of RNA transcription in prokaryotic organisms . As opposed to eukaryotes, who utilize a variety of proteins to initiate gene transcription, prokaryotic transcription is initiated almost completely by a σ-factor. The large and biologically essential protein, RNA polymerase (RNAP), contains one σ-subunit, which binds , located upstream of transcription start sites. Specific Function and StructureThe σ-factor performs two chief functions: to direct the catalytic core of RNAP to the +1 start site of transcription, and finally to assist in the initiation of strand seperation of double-helical DNA, ultimately forming the transcription "bubble." Each gene promoter utilizes a specific promoter region about 40 bp upstream of the transcription start site, and therefore different σ-factors play a role in the regulation of different genes. This process, which includes association of the σ-factor with RNAP to recognize and open DNA at the promoter site, followed by dissociation of the σ to allow elongation, is referred to as the σ cycle. DomainsThere are many types of σ-subunits, and each recognizes a unique promoter sequence. Furthmore, each σ is composed of a variable number of structured domains. The simplest σ-factors have two domains, few have three, and others, called housekeeping σ-factors have 4 domains. Each of these domains has DNA-binding determinants, or motifs that recognize specific sequences and conformations in DNA. Most commonly, these recognized motifs occur at the -35 and -10 locations upstream of the +1 site. One such DNA binding motif, the helix-turn-helix motif , helps specifically recognize DNA promoters at both the -35 and -10 positions. This HTH motif, used by most σ-factors, maintains its specificity and accuracy by binding in the major groove of DNA, where it can interact with the base pairs in the DNA double-helix. In many prokaryotes, these portions of DNA maintain consensus adenosine and thymine sequences, such as . RestrictionNormally, σ-factor domains cannot bind to promoters. These domains usually are placed in very compacted positions relative to each other, a conformation that buries DNA-binding determinants. This type of restriction is called conformational restriction. Additionally, in housekeeping σs, a domain called the σ(1.1) stabilizes the compact conformation mentioned above, thereby preventing any promoter recognition. Transcription BubbleThe is fundamentally formed through the common housekeeping σ factors which unwind about 13 bp of duplex DNA in an ATP independent process. As such, σ factors require invariant basic and aromatic residues (Phe, Tyr, Trp) critical for this formation. The process of bubble formation begins at the -11 formation (usually A) and propogates to +1 site, through "base flipping" which interrupts the stacking interactions stabilizing the double helix conformation. As a result, -35 and -10 motif sequence recognition and strand separation are both coupled by the σ factor. Gene Regulation and Differentiation3D structures of sigma factorUpdated on 13-October-2014 Sigma factor1ku2 – TaSF region 1.2-3.1 – Thermus aquaticus Sigma factor complex with DNA3ugo, 3ugp - TaSF region 2 + DNA Sigma factor complex with protein3lev – TaSF residues 93-271 + antibody Sigma factor complex with protein and DNA1rio - TaSF region 4 + repression protein CI + DNA ReferencesFelklistov, Andrey, Brian D. Sharon, Seth A. Darst, and Carol A. Gross. "Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective." The Annual Review of Microbiology 68 (2014): 357-76. Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd ed. Hoboken, NJ: Wiley, 2008. Mooney, R. A., S. A. Darst, and R. Landick. "Sigma and RNA Polymerase: An On-again, Off-again Relationship?" Molecular Cell 20.3 (2005): 335-45.
| ||||||||||||