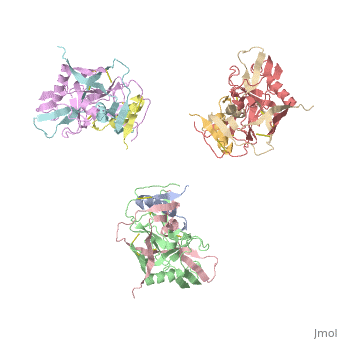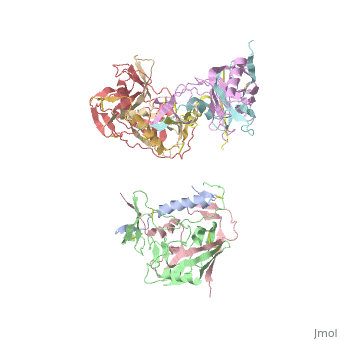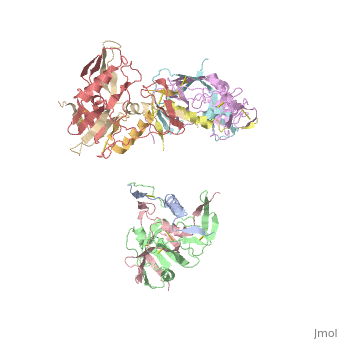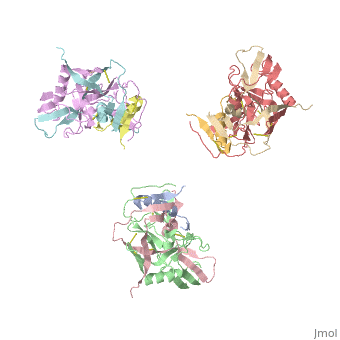Introduction
Human Immunodeficiency Virus (HIV) is a retrovirus/lentivirus that expresses a spike protein on its viral envelope (Env). "The envelope protein is synthesized as a precursor, gp160, which trimerizes and undergoes cleavage into two noncovalently associated fragments, the receptor-binding fragment gp120 and the fusion fragment gp41" [1]. Thus, Env is a heterodimer consisting of gp120 and gp41 and forms trimers on the surface of the viral membrane. As "the sole antigen on the virion surface," an understanding of Env (gp 120 & 41) and the conformational changes that occur in gp120 and gp41 during binding and fusion, respectively, is necessary in developing new fusion inhibitors and possible vaccines [1] [2]. The protein structure shown on the right is the gp120 portion of Env in its unbound trimer state [2].
Env Structure
The viral Env protein, as mentioned above, is composed of gp120 and gp41 that forms trimers on the viral membrane surface.
The image to the right, displays the undecorated Env trimer. Within the gp120 monomeric proteins are variable loops that undergo conformational changes that support binding or restrict binding of receptors such as CD4 or antibodies. Two important loops in this regard are the V1/V2 loops. The V1/V2 loops are located at the apex of the structure in the center of the three monomeric gp120 proteins. Depending on the gp120 conformation changes, the three gp41 transmembrane proteins, located beneath the gp120 surface proteins (located by the white arrow in F2.b), will undergo "irreversible refolding," bringing the viral membrane and host cell membrane closer together, consequently inducing fusion
[1].
gp120 Binding
gp120 is responsible for the binding of HIV to CD4 cells. "Binding of gp120 to the primary receptor CD4 and coreceptor (for example, CCR5 and CXCR4) induces large conformational changes, which then trigger dissociation of gp120 and a cascade of refolding events in gp41"
[1]. In the unliganded state, the are located at the center of the apex of the trimer and hold the trimer together. However, when binds to gp120, the V1/V2 loops undergo an outward rotational change that expose the central gp41 proteins at the base of the Env protein. gp120 is often complexed with and an antibody Fab (17b-Fab). "The 17b antibody has been shown to stabilize and lock gp120 in the CD4-bound conformation"
[2]. "Direct interatomic contacts are made between 22 CD4 residues and 26 gp120 amino-acid residues. These include 219 van der Waals contacts and 12 hydrogen bonds"
[3]. "Residues in contact are concentrated in the from 25 to 64 of CD4, but they are distributed over six segments of gp120"
[3]. However, the most important gp120/CD4 are between "Phe 43 and Arg 59 of CD4 [which] make multiple contacts centered on residues Asp 368, Glu 370 and Trp427 of gp120, which are all conserved among primate immunodeficiency viruses"
[3].
Broadly Neutralizing Antibody b12
b12 is a broadly neutralizing antibody that can bind to gp120 and inhibit the binding to primary corecptor CD4. "The binding of b12 results in a partial opening of the spike, coupled with roation of each monomer by ~20-25 degrees"[2]. Even though, b12 and CD4 bind roughly in the same areas, they shift the orientation of the gp120 monomers very differently. "Binding of the brodly neutralizing antibody b12... appears to lock gp120 and trimeric Env in a state that prevents further conformation changes that would normally lead to exposure of the gp41 stalk" [2].
Immune Evasion
One reason that HIV is able to escape inhibition by the immune system, is because of its conformational variability and resistance to antibodies. Essentially "the conformational flexibility of gp120 facilitates a decoy strategy that misdirects the humoral immune response" [4]. The interaction between CD4-binding site (CD4BS) antibodies and gp120 has been studied using many different antibodies. It has been observed that CD4BS antibodies displace the "four stranded 'bridging sheet'... to uncover a hydrophobic surface... that most CD4BS antibodies rely on access to" [4]. In the trimer this causes clashing between the gp120 monomers, which represent constraining factors that prevent the binding and inhibition by antibodies to gp120. The displacement of the bridging sheets, cause conformational changes in the V1/V2 causing clashes in the trimer. "Thus, CD4BS antibodies that access the hydrophobic region under the bridging sheet induce conformation in gp120 that are poorly compatible with the functional viral spike" [4]. "The bridging sheet and the V1/V2 loops detect and amplify any recognition that strays outside the target site" [4].