Sandbox viralpackagingmotors
From Proteopedia
This page is setup for Ojuewa to build her senior project for OU CHEM 4923
Contents |
Viral Packaging
|
One of the most important stage in the life cycle of all viruses is the encapsidation (packaging) of the viral genome. Many virus package their genome into preformed capsids using packaging motors powered by the hydrolysis of ATP. The hexameric ATPase P4 of dsRNA bacteriophage phi 12, located at the vertices of the icosahedral capsid is such a packaging motor. [1] P4 protein is a 35-kDa ssRNA packaging ATPase from dsRNA bacteriophages belonging to the Cystoviridae family. ( phi 6 - phi 12). These viruses use a packaging motor like RNA packaging motor that is powered by the hydrolysis of ATP to condense the nucleic acid into a confined space.
Structure
The RNA packaging motor is a straightforward machinery that consists of a portal protein (P4) hexamer. The crystal structure of the hexameric P4 protein is made of three sub-units, one-half of the hexameric molecule (hexamers encirlce the crystallographic 2-fold axes). Residues 196-206, 300-306, and the C-terminal segment 322-331are disordered in all subunits.
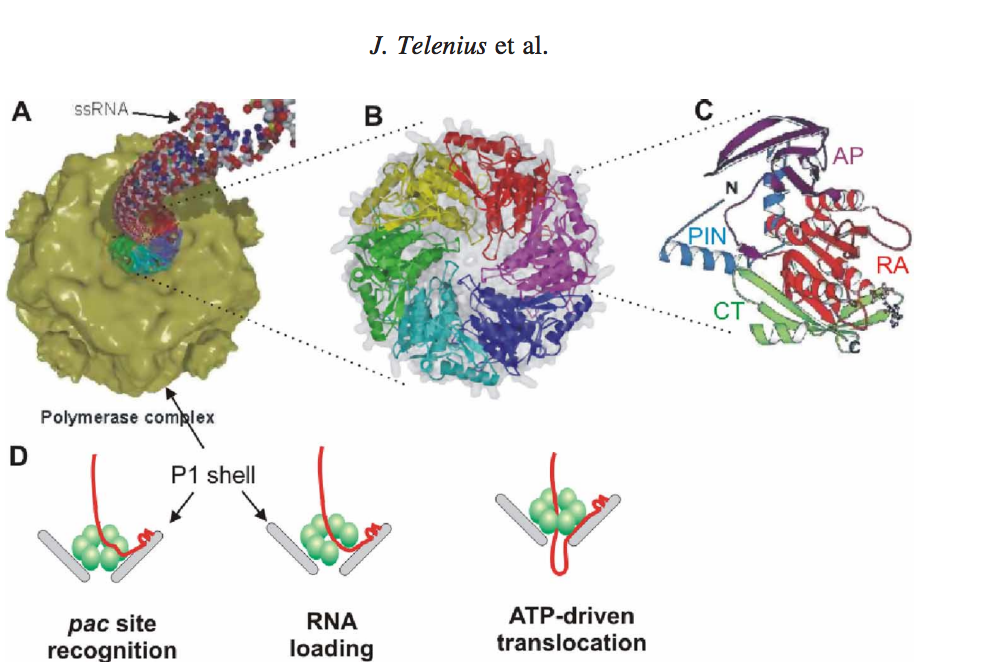 (A) Packaging of ssRNA via P4 hexamer into the icosahedral procapsid (B) P4 hexamer structure (C) Subunit structure, RecA-like catalytic core is shown in red (RA), N-terminal safety pin in cyan (PIN), apical domain in magenta (AP) and C-terminal platform in green (CT). The safety pin stabilizes interacts with a neighboring subunit and stabilizes the hexamer. (D) A scheme of the three step packaging mechanism for a ssRNA precursor. [2]
The P4 viral packaging motor is a sequential is a molecular machine that must complete several task in sequential order.
(A) Packaging of ssRNA via P4 hexamer into the icosahedral procapsid (B) P4 hexamer structure (C) Subunit structure, RecA-like catalytic core is shown in red (RA), N-terminal safety pin in cyan (PIN), apical domain in magenta (AP) and C-terminal platform in green (CT). The safety pin stabilizes interacts with a neighboring subunit and stabilizes the hexamer. (D) A scheme of the three step packaging mechanism for a ssRNA precursor. [2]
The P4 viral packaging motor is a sequential is a molecular machine that must complete several task in sequential order.
Function
The RNA packaging motor is a straightforward machinery that consists of a portal protein (P4) hexamer.
Energetics
The P4 protein provides energy for the RNA translocation.
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
- ↑ Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004 Sep 17;118(6):743-55. PMID:15369673 doi:10.1016/j.cell.2004.09.007
- ↑ http://dx.doi.org/10.1080/17486700802168502